Molecular and clinical characterization of Colombian patients suffering from type III glycogen storage disease
Abstract
Introduction: Type III glycogen storage disease (GSD III) is an autosomal recessive disorder in which a mutation in the AGL gene causes deficiency of the glycogen debranching enzyme. The disease is characterized by fasting hypoglycemia, hepatomegaly and progressive myopathy. Molecular analyses of AGL have indicated heterogeneity depending on ethnic groups. The full spectrum of AGL mutations in Colombia remains unclear.
Objective: To describe the clinical and molecular characteristics of ten Colombian patients diagnosed with GSD III.
Materials and methods: We recruited ten Colombian children with a clinical and biochemical diagnosis of GSD III to undergo genetic testing. The full coding exons and the relevant exon-intron boundaries of the AGL underwent Sanger sequencing to identify mutation.
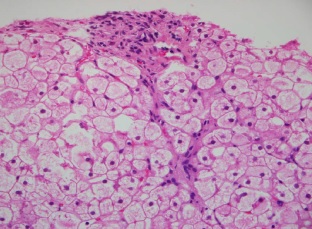
Results: All patients had the classic phenotype of the GSD III. Genetic analysis revealed a mutation p.Arg910X in two patients. One patient had the mutation p.Glu1072AspfsX36, and one case showed a compound heterozygosity with p.Arg910X and p.Glu1072AspfsX36 mutations. We also detected the deletion of AGL gene 3, 4, 5, and 6 exons in three patients. The in silico studies predicted that these defects are pathogenic. No mutations were detected in the amplified regions in three patients.
Conclusion: We found mutations and deletions that explain the clinical phenotype of GSD III patients. This is the first report with a description of the clinical phenotype and the spectrum of AGL mutations in Colombian patients. This is important to provide appropriate prognosis and genetic counseling to the patient and their relatives.
Downloads
References
Wolfsdorf JI, Weinstein DA. Glycogen storage diseases. Rev Endocr Metab Disord. 2003;4:95-102.
Shin YS. Glycogen storage disease: Clinical, biochemical, and molecular heterogeneity. Semin Pediatr Neurol. 2006;13:115-20. https://doi.org/10.1016/j.spen.2006.06.007
Ozen H. Glycogen storage diseases: New perspectives. World J Gastroenterol. 2007;13:2541-53. https://doi.org/10.3748/wjg.v13.i18.2541
Cheng A, Zhang M, Okubo M, Omichi K, Saltiel AR. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet. 2009;18:2045-52. https://doi.org/10.1093/hmg/ddp128
Endo Y, Fateen E, Aoyama Y, Horinishi A, Ebara T, Murase T, et al. Molecular characterization of Egyptian patients with glycogen storage disease type IIIa. J Hum Genet. 2005;50:538-42. https://doi.org/10.1007/s10038-005-0291-3
Shen JJ, Chen YT. Molecular characterization of glycogen storage disease type III. Curr Mol Med. 2002;2:167-75. https://doi.org/10.2174/1566524024605752
Haagsma EB, Smit GP, Niezen-Koning KE, Gouw AS, Meerman L, Slooff MJ. Type IIIb glycogen storage disease associated with end-stage cirrhosis and hepatocellular carcinoma. The Liver Transplant Group. Hepatology. 1997;25:537-40. https://doi.org/10.1002/hep.510250307
Sentner CP, Hoogeveen IJ, Weinstein DA, Santer R, Murphy E, McKiernan PJ, et al. Glycogen storage disease type III: Diagnosis, genotype, management, clinical course and outcome. J Inherit Metab Dis. 2016;39:697-704. https://doi.org/10.1007/s10545-016-9932-2
Rousseau-Nepton I, Okubo M, Grabs R, Consortium FC, Mitchell J, Polychronakos C, et al. A founder AGL mutation causing glycogen storage disease type IIIa in Inuit identified through whole-exome sequencing: A case series. CMAJ. 2015;187:E68-73. https://doi.org/10.1503/cmaj.140840
Mili A, Ben Charfeddine I, Mamai O, Abdelhak S, Adala L, Amara A, et al. Molecular and biochemical characterization of Tunisian patients with glycogen storage disease type III. J Hum Genet. 2012;57:170-5. https://doi.org/10.1038/jhg.2011.122
Parvari R, Moses S, Shen J, Hershkovitz E, Lerner A, Chen YT. A single-base deletion in the 3’-coding region of glycogen-debranching enzyme is prevalent in glycogen storage disease type IIIA in a population of North African Jewish patients. Eur J Hum Genet. 1997;5:266-70.
Santer R, Kinner M, Steuerwald U, Kjaergaard S, Skovby F, Simonsen H, et al. Molecular genetic basis and prevalence of glycogen storage disease type IIIA in the Faroe Islands. Eur J Hum Genet. 2001;9:388-91. https://doi.org/10.1038/sj.ejhg.5200632
Talente GM, Coleman RA, Alter C, Baker L, Brown BI, Cannon RA, et al. Glycogen storage disease in adults. Ann Intern Med. 1994;120:218-26.
Lucchiari S, Santoro D, Pagliarani S, Comi GP. Clinical, biochemical and genetic features of glycogen debranching enzyme deficiency. Acta Myol. 2007;26:72-4.
van Hoof F, Hers HG. The subgroups of type 3 glycogenosis. Eur J Biochem. 1967;2:265-70. https://doi.org/10.1111/j.1432-1033.1967.tb00134.x
Ding JH, de Barsy T, Brown BI, Coleman RA, Chen YT. Immunoblot analyses of glycogen debranching enzyme in different subtypes of glycogen storage disease type III. J Pediatr. 1990;116:95-100.
Yang-Feng TL, Zheng K, Yu J, Yang BZ, Chen YT, Kao FT. Assignment of the human glycogen debrancher gene to chromosome 1p21. Genomics. 1992;13:931-4. https://doi.org/10.1016/0888-7543(92)90003-B
Yang BZ, Ding JH, Enghild JJ, Bao Y, Chen YT. Molecular cloning and nucleotide sequence of cDNA encoding human muscle glycogen debranching enzyme. J Biol Chem. 1992;267:9294-9.
Bao Y, Yang BZ, Dawson TL Jr, Chen YT. Isolation and nucleotide sequence of human liver glycogen debranching enzyme mRNA: Identification of multiple tissue-specific isoforms. Gene. 1997;197:389-98. https://doi.org/10.1016/S0378-1119(97)00291-6
Lucchiari S, Fogh I, Prelle A, Parini R, Bresolin N, Melis D, et al. Clinical and genetic variability of glycogen storage disease type IIIa: Seven novel AGL gene mutations in the Mediterranean area. Am J Med Genet. 2002;109:183-90. https://doi.org/10.1002/ajmg.10347
Horinishi A, Okubo M, Tang NL, Hui J, To KF, Mabuchi T, et al. Mutational and haplotype analysis of AGL in patients with glycogen storage disease type III. J Hum Genet. 2002;47:55-9. https://doi.org/10.1007/s100380200000
Okubo M, Horinishi A, Suzuki Y, Murase T, Hayasaka K. Compound heterozygous patient with glycogen storage disease type III: Identification of two novel AGL mutations, a donor splice site mutation of Chinese origin and a 1-bp deletion of Japanese origin. Am J Med Genet. 2000;93:211-4. https://doi.org/10.1016/S0378-1119(97)00291-6
Okubo M, Horinishi A, Takeuchi M, Suzuki Y, Sakura N, Hasegawa Y, et al. Heterogeneous mutations in the glycogen-debranching enzyme gene are responsible for glycogen storage disease type IIIa in Japan. Hum Genet. 2000;106:108-15.
Ogimoto A, Okubo M, Okayama H, Shin YS, Endo Y, Ebara T, et al. A Japanese patient with cardiomyopathy caused by a novel mutation R285X in the AGL gene. Circ J. 2007;71:1653-6. https://doi.org/10.1253/circj.71.1653
Endo Y, Fateen E, El Shabrawy M, Aoyama Y, Ebara T, Murase T, et al. Egyptian glycogen storage disease type III - identification of six novel AGL mutations, including a large 1.5 kb deletion and a missense mutation p.L620P with subtype IIId. Clin Chem Lab Med. 2009;47:1233-8. https://doi.org/10.1515/CCLM.2009.281
Goldstein JL, Austin SL, Boyette K, Kanaly A, Veerapandiyan A, Rehder C, et al. Molecular analysis of the AGL gene: Identification of 25 novel mutations and evidence of genetic heterogeneity in patients with glycogen storage disease type III. Genet Med. 2010;12:424-30. https://doi.org/10.1097/GIM.0b013e3181d94eaa
Aoyama Y, Ozer I, Demirkol M, Ebara T, Murase T, Podskarbi T, et al. Molecular features of 23 patients with glycogen storage disease type III in Turkey: A novel mutation p.R1147G associated with isolated glucosidase deficiency, along with 9 AGL mutations. J Hum Genet. 2009;54:681-6. https://doi.org/10.1038/jhg.2009.100
Sentner CP, Vos YJ, Niezen-Koning KN, Mol B, Smit GP. Mutation analysis in glycogen storage disease type III patients in the Netherlands: Novel genotype-phenotype relationships and five novel mutations in the AGL gene. JIMD Rep. 2013;7:19-26. https://doi.org/10.1007/8904_2012_134
Zimmermann A, Rossmann H, Bucerzan S, Grigorescu-Sido P. A novel nonsense mutation of the AGL gene in a Romanian patient with glycogen storage disease type IIIa. Case Rep Genet. 2016;2016:8154910. https://doi.org/10.1155/2016/8154910
Rhouma FB, Messai H, Hsouna S, Halim NB, Cherif W, Fadhel SB, et al. History of settlement of villages from Central Tunisia by studying families sharing a common founder glycogenosis type III mutation. Mitochondrial DNA A DNA MappSeq Anal. 2016;27:3194-8. https://doi.org/10.3109/19401736.2015.1007331
Lu C, Qiu Z, Sun M, Wang W, Wei M, Zhang X. Spectrum of AGL mutations in Chinese patients with glycogen storage disease type III: Identification of 31 novel mutations. J Hum Genet. 2016;61:641-5. https://doi.org/10.1038/jhg.2016.24
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073-81. https://doi.org/10.1038/nprot.2009.86
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;7:20. https://doi.org/10.1002/0471142905.hg0720s76
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310-5. https://doi.org/10.1038/ng.2892
den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS Recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564-9. https://doi.org/10.1002/humu.22981
den Dunnen JT. Sequence variant descriptions: HGVS nomenclature and mutalyzer. Curr Protoc Hum Genet. 2016;90:7.13.1-7.13.19. https://doi.org/10.1002/cphg.2
Aradhya S, Lewis R, Bonaga T, Nwokekeh N, Stafford A, Boggs B, et al. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet Med. 2012;14:594-603. https://doi.org/10.1038/gim.2011.65
Retterer K, Scuffins J, Schmidt D, Lewis R, Pineda Álvarez D, Stafford A, et al. Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. Genet Med. 2015;17:623-9. https://doi.org/10.1038/gim.2014.160
Dagli A, Sentner CP, Weinstein DA. Glycogen storage disease type III. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, et al., editors. Gene Reviews. Washington, D.C.: University of Washington; 1993.
Kishnani PS, Austin SL, Arn P, Bali DS, Boney A, Case LE, et al. Glycogen storage disease type III diagnosis and management guidelines. Genet Med. 2010;12:446-63.
https://doi.org/10.1097/GIM.0b013e3181e655b6
Coleman RA, Winter HS, Wolf B, Gilchrist JM, Chen YT. Glycogen storage disease type III (glycogen debranching enzyme deficiency): Correlation of biochemical defects with myopathy and cardiomyopathy. Ann Intern Med. 1992;116:896-900. https://doi.org/10.7326/0003-4819-116-11-896
Maire I, Baussan C, Moatti N, Mathieu M, Lemonnier A. Biochemical diagnosis of hepatic glycogen storage diseases: 20 years French experience. Clin Biochem. 1991;24:169-78. https://doi.org/10.1016/0009-9120(91)90511-C
Lucchiari S, Pagliarani S, Salani S, Filocamo M, Di Rocco M, Melis D, et al. Hepatic and neuromuscular forms of glycogenosis type III: Nine mutations in AGL. Hum Mutat. 2006;27:600-1. https://doi.org/10.1002/humu.9426
Lucchiari S, Donati MA, Melis D, Filocamo M, Parini R, Bresolin N, et al. Mutational analysis of the AGL gene: Five novel mutations in GSD III patients. Hum Mutat. 2003;22:337. https://doi.org/10.1002/humu.9177
Zhang W, Cui H, Wong LJ. Application of next generation sequencing to molecular diagnosis of inherited diseases. Top Curr Chem. 2014;336:19-45. https://doi.org/10.1007/128_2012_325
Yubero D, Brandi N, Ormazabal A, García-Cazorla A, Pérez-Dueñas B, Campistol J, et al. Targeted next generation sequencing in patients with inborn errors of metabolism. PLoS One. 2016;11:e0156359. https://doi.org/10.1371/journal.pone.0156359
Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: To decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499-505.
https://doi.org/10.1016/j.febslet.2008.12.058
Horinishi A, Murase T, Okubo M. Novel intronic polymorphisms (IVS6-73A/G and IVS21+124A/G) in the glycogen-debranching enzyme (AGL) gene. Hum Mutat. 2000;16:279. https://doi.org/10.1002/1098-1004(200009)16:
<279::AID-HUMU32>3.0.CO;2-X
Shamseldin HE, Al-Dosari M, Al-Jbali L, Rahbeeni Z, Qari A, Hashem M, et al. Study of consanguineous populations can improve the annotation of SNP databases. Eur J Med Genet. 2011;54:118-20. https://doi.org/10.1016/j.ejmg.2010.10.009
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























