Mutations in the Helicobacter pylori 23S rRNA gene associated with clarithromycin resistance in patients at an endoscopy unit in Medellín, Colombia
Abstract
Introduction: Clarithromycin is the first-line antibiotic for the treatment of Helicobacter pylori infection. Bacterial resistance is mainly due to the presence of specific mutations in the 23S ribosomal RNA (rRNA) gene.
Objective: To determine the frequency of A2143G and A2142G specific mutations in the 23S rRNA gene associated with clarithromycin resistance of H. pylori in samples from patients with dyspeptic manifestations in Medellín, northwestern Colombia.
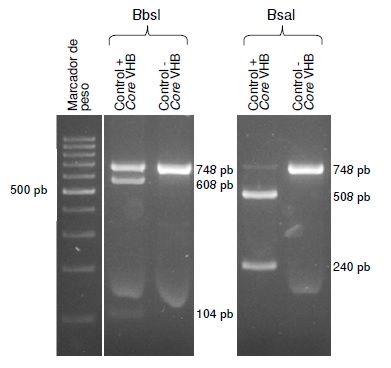
Materials and methods: DNA was extracted from gastric biopsy samples of patients with dyspeptic manifestations seen at an endoscopy unit in Medellín between 2016 and 2017. PCR was performed to amplify the bacterial s and m vacA regions, and a region in the 23S rRNA gene. The presence of the A2142G and A2143G mutations was determined using the restriction fragment length polymorphism (RFLP) technique with the BbsI and BsaI enzymes, respectively.
Results: The prevalence of infection was 44.2% (175/396), according to the histopathology report. The positive samples were analyzed and the three regions of the bacterial genome were amplified in 143 of the 175 samples. The A2143G and A2142G mutations were identified in 27 samples (18.8%, 27/143). The most frequent mutation was A2143G (81.5%, 22/27).
Conclusions: We found a high prevalence of H. pylori mutations associated with clarithromycin resistance in the study population. Further studies are required to determine the bacterial resistance in the Colombian population in order to define first line and rescue treatments.
Downloads
References
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. IARC monographs on the evaluation of carcinogenic risks to humans. Volume 100. A review of human carcinogens. Part B. Biological agents. Lyon: International Agency for Research on Cancer; 2012. p. 385-435.
Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39:14-23. https://doi.org/10.1016/j.bj.2015.06.002
Zabala B, Lucero Y, Lagomarcino AJ, Orellana-Manzano A, George S, Torres JP, et al. Prevalence and dynamics of Helicobacter pylori infection during childhood. Helicobacter. 2017;22:e12399. https://doi.org/10.1111/hel.12399
Hunt RH, Xiao SD, Megraud F, León-Barua R, Bazzoli F, van Der Merwe S, et al. Helicobacter pylori in developing countries. World Gastroenterology Organisation Global Guideline. J Gastrointestin Liver Dis. 2011;20:299-304.
Hooi JK, Lai WY, Ng WK, Suen MM, Underwood FE, Tanyingoh D, et al. Global prevalence of Helicobacter pylori Infection: Systematic review and meta-analysis. Gastroenterology. 2017;153:420-9. https://doi.org/10.1053/j.gastro.2017.04.022
Wroblewski LE, Peek RM, Wilson KT, Wilson KT. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713-39. https://doi.org/10.1128/CMR.00011-10
Gisbert JP. Enfermedades relacionadas con la infección por Helicobacter pylori. Gastroenterol Hepatol. 2013;36:39-50.
Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC Cancerbase No. 11. Lyon: International Agency for Research on Cancer; 2013.
Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. https://doi.org/10.1136/gutjnl-2016-312288
Martínez JD, Henao SC, Lizarazo JI. Antibiotic resistance of Helicobacter pylori in Latin America and the Caribbean. Rev Colomb Gastroenterol. 2014;29:218-27.
Versalovic J, Osato MS, Spakovsky K, Dore MP, Reddy R, Stone GG, et al. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283-6.
Trespalacios AA, Rimbara E, Otero W, Reddy R, Graham DY. Improved allele-specific PCR assays for detection of clarithromycin and fluoroquinolone resistant of Helicobacter pylori in gastric biopsies: Identification of N87I mutation in GyrA. Diagn Microbiol Infect Dis. 2015;81:251-5. https://doi.org/10.1016/j.diagmicrobio.2014.12.003
Ciftci IH, Ugras M, Acartekin G, Asik G, Safak B, Dilek FH. Comparison of FISH, RFLP and agar dilution methods for testing clarithromycin resistance of Helicobacter pylori. Turkish J Gastroenterol. 2015;25:75-80. https://doi.org/10.5152/tjg.2014.4688
Trespalacios AA, Otero W, Caminos JE, Mercado MM, Ávila J, Rosero LE, et al. Phenotypic and genotypic analysis of clarithromycin-resistant Helicobacter pylori from Bogotá, D.C., Colombia. J Microbiol. 2013;51:448-52. https://doi.org/10.1007/s12275-013-2465-6
Kim KS, Kang JO, Eun CS, Han DS, Choi TY. Mutations in the 23S rRNA gene of Helicobacter pylori associated with clarithromycin resistance. J Korean Med Sci. 2002;17:599-603. https://doi.org/10.3346/jkms.2002.17.5.599
Trespalacios AA, Otero-Regino W, Mercado-Reyes M. Helicobacter pylori resistance to metronidazole, clarithromycin and amoxicillin in Colombian patients. Rev Colomb Gastroenterol. 2010;25:29-36.
Bravo LE, Cortés A, Carrascal E, Jaramillo R, García LS, Bravo PE, et al. Helicobacter pylori: patología y prevalencia en biopsias gástricas en Colombia. Colomb Med. 2003;34:124-31.
Lee YC, Liou JM, Wu MS, Wu CY, Lin JT. Eradication of Helicobacter pylori to prevent gastroduodenal diseases: Hitting more than one bird with the same stone. Therap Adv Gastroenterol. 2008;1:111-20. https://doi.org/10.1177/1756283X08094880
Instituto Nacional de Salud. Quinto Informe ONS: carga de enfermedad por enfermedades crónicas no transmisibles y discapacidad en Colombia. Quinta edición. Bogotá: Instituto Nacional de Salud; 2015.
Regino WO. La importancia de cultivar Helicobacter pylori. Rev Colomb Gastroenterol. 2013;28:87-92.
Sierra F, Forero JD, Rey M. Tratamiento ideal del Helicobacter pylori: una revisión sistemática. Rev Gastroenterol Mex. 2014;79:28-49. https://doi.org/10.1016/j.rgmx.2013.03.003
Gómez M, Ruíz O, Páramo-Hernández D, Albis R, Sabbagh LC. Erradicación del Helicobacter pylori: encuesta realizada por la Asociación Colombiana de Gastroenterología. Rev Col Gastroenterol. 2015;30:25-31.
Camargo MC, García A, Riquelme A, Otero W, Camargo CA, Hernández-García T, et al. The problem of Helicobacter pylori resistance to antibiotics: A systematic review in Latin America. Am J Gastroenterol. 2014;109:485-95. https://doi.org/10.1038/ajg.2014.24
Di Ciaula A, Scaccianoce G, Venerito M, Zullo A, Bonfrate L, Rokkas T, et al. Eradication rates in italian subjects heterogeneously managed for Helicobacter pylori infection. Time to abandon empiric treatments in Southern Europe. J Gastrointest Liver Dis. 2017;26:129-37. https://doi.org/10.15403/jgld.2014.1121.262.itl
Castaño R, Ruiz-Vélez MH, Martínez-Hincapié C, Naranjo-Aristizábal FA, Campuzano-Maya G, Sanín-Fonnegra E, et al. Evaluación para comparar dos esquemas de terapia estándar (7 frente a 10 días) contra el Helicobacter pylori, con seguimiento clínico a 1 año. Rev Colomb Gastroenterol. 2012;27:80-7.
Castaño R, Ruiz-Vélez MH, Campuzano-Maya G, Sanín-Fonnegra E, Puerta-Díaz JD, Calvo-Betancur V, et al. Randomized study comparing standard first line 10 day therapy against Helicobacter pylori including clarithromycin versus standard first line therapy with levofloxacin. Rev Colomb Gastroenterol. 2013;28:101-8.
Correa S, Cardona AF, Correa T, Correa LA, García HI, Estrada S. Prevalence of Helicobacter pylori and histopathological features in gastric biopsies from patients with dyspeptic symptoms at a referral center in Medellín. Rev Colomb Gastroenterol. 2016;31:9-15.
Peixoto A, Silva M, Pereira P, Macedo G. Biopsies in gastrointestinal endoscopy: When and how. GE Port J Gastroenterol. 2016;23:19-27. https://doi.org/10.1016/j.jpge.2015.07.004
Genta RM, Graham DY. Comparison of biopsy sites for the histopathologic diagnosis of Helicobacter pylori: A topographic study of H. pylori density and distribution. Gastrointest Endosc. 1994;40:342-5. https://doi.org/10.1016/S0016-5107(94)70067-2
Acosta CP, Hurtado FA, Trespalacios AA. Determinación de mutaciones de un solo nucleótido en el gen 23S rRNA de Helicobacter pylori relacionadas con resistencia a claritromicina en una población del departamento del Cauca, Colombia. Biomédica. 2013;34:156-62. https://doi.org/10.7705/biomedica.v34i0.1649
Occhialini A, Urdaci M, Doucet-Populaire F, Bébéar CM, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: Rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724-8. https://doi.org/10.1128/AAC.41.12.2724
Rendón JC, Cortés-Mancera F, Restrepo-Gutiérrez JC, Hoyos S, Navas MC. Molecular characterization of occult hepatitis B virus infection in patients with end-stage liver disease in Colombia. PLoS One. 2017;12:e0180447. https://doi.org/10.1371/journal.pone.0180447
Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: What should be the gold standard? World J Gastroenterol. 2014;20:12847-59. https://doi.org/10.3748/wjg.v20.i36.12847
Jiménez F, Barbaglia Y, Bucci P, Tedeschi FA, Zalazar FE. Detección molecular y genotipificación de Helicobacter pylori en biopsias gástricas de pacientes adultos sintomáticos de la ciudad de Santa Fe, Argentina. Rev Argent Microbiol. 2013;45:39-43.
Wang YK, Kuo FC, Liu CJ, Wu MC, Shih HY, Wang SS, et al. Diagnosis of Helicobacter pylori infection: Current options and developments. World J Gastroenterol. 2015;21:11221-35. https://doi.org/10.3748/wjg.v21.i40.11221
de Martel C, Plummer M, van Doorn LJ, Vivas J, López G, Carillo E, et al. Comparison of polymerase chain reaction and histopathology for the detection of Helicobacter pylori in gastric biopsies. Int J Cancer. 2010;126:1992-6. https://doi.org/10.1002/ijc.24898
Cosgun Y, Yildirim A, Yucel M, Karakoc AE, Koca G, Gonultas A, et al. Evaluation of invasive and noninvasive methods for the diagnosis of Helicobacter pylori infection. Asian Pac J Cancer Prev. 2016;17:6165-72. https://doi.org/10.22034/APJCP.2016.17.12.5265
Hammar M, Tyszkiewicz T, Wadström T, O’Toole PW. Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J Clin Microbiol. 1992;30:54-8.
Ahmad N, Zakaria WR, Abdullah SA, Mohamed R. Characterization of clarithromycin resistance in Malaysian isolates of Helicobacter pylori. World J Gastroenterol. 2009;15:3161-5. https://doi.org/10.3748/wjg.15.3161
Yula E, Nagiyev T, Kaya ÖA, Inci M, Çelik MM, Köksal F. Detection of primary clarithromycin resistance of Helicobacter pylori and association between cagA + status and clinical outcome. Folia Microbiol (Praha). 2013;58:141-6. https://doi.org/10.1007/s12223-012-0192-8
Mahmoudi S, Mamishi S, Banar M, Keshavarz Valian S, Bahador A, Najafi M, et al. Antibiotic susceptibility of Helicobacter pylori strains isolated from Iranian children: High frequency of A2143G point mutation associated with clarithromycin resistance. J Glob Antimicrob Resist. 2017;10:131-5. https://doi.org/10.1016/j.jgar.2017.04.011
Abdollahi H, Savari M, Zahedi MJ, Moghadam SD, Abasi MH. Detection of A2142C, A2142G, and A2143G mutations in 23s rRNA gene conferring resistance to clarithromycin among Helicobacter pylori isolates in Kerman, Iran. Iran J Med Sci. 2011;36:104-10.
Pourakbari B, Mahmoudi S, Parhiz J, Sadeghi RH, Monajemzadeh M, Mamishi S. High frequency of metronidazole and clarithromycin-resistant Helicobacter pylori in formalin-fixed, paraffin-embedded gastric biopsies. Br J Biomed Sci. 2018;75:61-5. https://doi.org/10.1080/09674845.2017.1391466
Henao SC, Quiroga A, Martínez JD, Otero W. Resistencia primaria a la claritromicina en aislamientos de Helicobacter pylori. Rev Colomb Gastroenterol. 2009;24:110-4.
Figueroa M, Cortés A, Pazos A, Bravo LE. Susceptibilidad in vitro de Helicobacter pylori a amoxicilina y claritromicina obtenido a partir de biopsias gástricas de pacientes de zona de bajo riesgo para cáncer gástrico. Biomédica. 2012;32:32-42. https://doi.org/10.7705/biomedica.v32i1.454
Yepes CA, Rodríguez A, Ruiz Á, Ariza B. Antibiotics resistance of Helicobacter pylori at the San Ignacio University Hospital in Bogotá. Acta Medica Colomb. 2008;33:11-4.
Álvarez A, Moncayo JI, Santacruz JJ, Corredor LF, Reinosa E, Martínez JW, et al. Resistencia a metronidazol y claritromicina en aislamientos de Helicobacter pylori de pacientes dispépticos en Colombia. Rev Med Chil. 2009;137:1309-14. https://doi.org/10.4067/S0034-98872009001000005
Álvarez A, Moncayo JI, Santacruz JJ, Santacoloma M, Corredor LF, Reinosa E. Antimicrobial susceptibility and mutations involved in clarithromycin resistance in Helicobacter pylori isolates from patients in the western central region of Colombia. Antimicrob Agents Chemother. 2009;53:4022-4. https://doi.org/10.1128/AAC.00145-09
Bustamante-Rengifo JA, Matta AJ, Pazos A, Bravo LE. In vitro effect of amoxicillin and clarithromycin on the 3’ region of cagA gene in Helicobacter pylori isolates. World J Gastroenterol. 2013;19:6044-54. https://doi.org/10.3748/wjg.v19.i36.6044
Sanches BS, Martins GM, Lima K, Cota B, Moretzsohn LD, Ribeiro LT, et al. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: A national survey observational study. World J Gastroenterol. 2016;22:7587-94. https://doi.org/10.3748/wjg.v22.i33.7587
Zerbetto G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, et al. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microb Drug Resist. 2016;23:351-8. https://doi.org/10.1089/mdr.2015.0361
Alarcón-Millán J, Fernández-Tilapa G, Cortés-Malagón EM, Castañón-Sánchez CA, De Sampedro-Reyes J, Cruz-del Carmen I, et al. Clarithromycin resistance and prevalence of Helicobacter pylori virulent genotypes in patients from Southern México with chronic gastritis. Infect Genet Evol. 2016;44:190-8. https://doi.org/10.1016/j.meegid.2016.06.044
Trespalacios-Rangel AA, Otero W, Arévalo-Galvis A, Poutou-Piñales RA, Rimbara E, Graham DY. Surveillance of levofloxacin resistance in Helicobacter pylori isolates in Bogotá-Colombia (2009-2014). PLoS One. 2016;11:e0160007. https://doi.org/10.1371/journal.pone.0160007
Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE, et al. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43:514-33. https://doi.org/10.1111/apt.13497
Some similar items:
- Mercedes Figueroa, Armando Cortés, Álvaro Pazos, Luis Eduardo Bravo, Antimicrobial susceptibility of Helicobacter pylori with chronic gastritis , Biomedica: Vol. 32 No. 1 (2012)
- Claudia Patricia Acosta, Fabián Andrés Hurtado, Alba Alicia Trespalacios, Determination of single nucleotide mutations in the 23S rRNA gene of Helicobacter pylori related to clarithromycin resistance in a population from Cauca, Colombia , Biomedica: Vol. 34 (2014): Abril, Suplemento 1, Resistencia bacteriana
- Yeison Harvey Carlosama , Claudia Patricia Acosta , Carlos Hernán Sierra , Carol Yovanna Rosero , Harold Jofre Bolaños, The Operative Link on Gastritis Assessment (OLGA) system as a marker for gastric cancer and dysplasia in a Colombian population at risk: A multicenter study , Biomedica: Vol. 43 No. Sp. 3 (2023): Enfermedades crónicas no transmisibles
- Juan Camilo Caguazango, Álvaro Jairo Pazos, Microbiota according to gastric topography in patients with low or high risk of gastric cancer in Nariño, Colombia , Biomedica: Vol. 39 No. Supl. 2 (2019): Enfermedades transmisibles en el trópico, agosto
- María Teresa Arango, Carlos Jaramillo, María Camila Montealegre, Mabel Helena Bohórquez, María del Pilar Delgado, Genetic characterization of the interleukin 1 β polymorphisms -511, -31 y +3954 in a Colombian population with dyspepsia , Biomedica: Vol. 30 No. 2 (2010)
- Andrés Javier Quiroga, Antonio Huertas, Alba Lucía Cómbita, María Mercedes Bravo, Variation in the number of EPIYA-C repeats in CagA protein from Colombian Helicobacter pylori strains and its ability to induce hummingbird phenotype in gastric epithelial cells , Biomedica: Vol. 30 No. 2 (2010)
- Lino E. Torres, Lidice González, Karelia Melián, Jordis Alonso, Arlenis Moreno, Mayrín Hernández, Orlando Reyes, Ludisleydis Bermúdez, Javier Campos, Guillermo Pérez-Pérez, Boris L. Rodríguez, EPIYA motif patterns among Cuban Helicobacter pylori CagA positive strains , Biomedica: Vol. 32 No. 1 (2012)
- Andrés Javier Quiroga, Diana Marcela Cittelly, María Mercedes Bravo, BabA2, oipA and cagE Helicobacter pylori genotypes in Colombian patients with gastroduodenal diseases. , Biomedica: Vol. 25 No. 3 (2005)
- Lidice González, Karen Marrero, Orlando Reyes, Elaine Rodríguez, Liudmila Martínez, Boris L. Rodríguez, Cloning and expression of a recombinant CagA-gene fragment of Helicobacter pylori and its preliminary evaluation in serodiagnosis , Biomedica: Vol. 33 No. 4 (2013)
- Esperanza Trujillo, Teresa Martínez, María Mercedes Bravo, Genotyping of Helicobacter pylori virulence factors vacA and cagA in individuals from two regions in Colombia with opposing risk for gastric cancer , Biomedica: Vol. 34 No. 4 (2014)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























