Isolation and characterization of an early SARS-CoV-2 isolate from the 2020 epidemic in Medellín, Colombia
Abstract
Introduction: SARS-CoV-2 has been identified as the new coronavirus causing an outbreak of acute respiratory disease in China in December, 2019. This disease, currently named COVID-19, has been declared as a pandemic by the World Health Organization (WHO). The first case of COVID-19 in Colombia was reported on March 6, 2020. Here we characterize an early SARS-CoV-2 isolate from the pandemic recovered in April, 2020.
Objective: To describe the isolation and characterization of an early SARS-CoV-2 isolate from the epidemic in Colombia.
Materials and methods: A nasopharyngeal specimen from a COVID-19 positive patient was inoculated on different cell lines. To confirm the presence of SARS-CoV-2 on cultures we used qRT-PCR, indirect immunofluorescence assay, transmission and scanning electron microscopy, and next-generation sequencing.
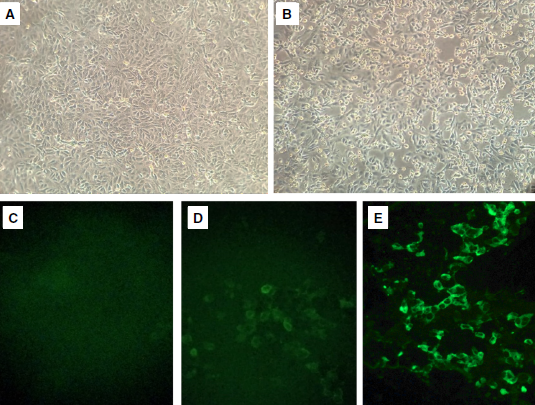
Results: We determined the isolation of SARS-CoV-2 in Vero-E6 cells by the appearance of the cytopathic effect three days post-infection and confirmed it by the positive results in the qRT-PCR and the immunofluorescence with convalescent serum. Transmission and scanning electron microscopy images obtained from infected cells showed the presence of structures compatible with SARS-CoV-2. Finally, a complete genome sequence obtained by next-generation sequencing allowed classifying the isolate as B.1.5 lineage.
Conclusion: The evidence presented in this article confirms the first isolation of SARSCoV-2 in Colombia. In addition, it shows that this strain behaves in cell culture in a similar way to that reported in the literature for other isolates and that its genetic composition is consistent with the predominant variant in the world. Finally, points out the importance of viral isolation for the detection of neutralizing antibodies, for the genotypic and phenotypic characterization of the strain and for testing compounds with antiviral potential.
Downloads
References
Huang D, Lian X, Song F, Ma H, Lian Z, Liang Y, et al. Clinical features of severe patients infected with 2019 novel coronavirus: A systematic review and meta-analysis. Ann Transl Med. 2020;8:576. https://doi.org/10.21037/atm-20-2124
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9. https://doi.org/10.1038/s41586-020-2008-3
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-3. https://doi.org/10.1038/s41586-020-2012-7
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-33. https://doi.org/10.1056/NEJMoa2001017
Bai Y, Jiang D, Lon JR, Chen X, Hu M, Lin S, et al. Comprehensive evolution and molecular characteristics of a large number of SARS-CoV-2 genomes reveal its epidemic trends. Int J Infect Dis. 2020;100:164-73. https://doi.org/10.1016/j.ijid.2020.08.066
Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495-504. https://doi.org/10.2471/BLT.20.253591
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812-27 e19. https://doi.org/10.1016/j.cell.2020.06.043
Toyoshima, Y., Nemoto, K., Matsumoto, S. et al. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020. https://doi.org/10.1038/s10038-020-0808-9
Ministerio de Salud y Protección Social. Colombia confirma su primer caso de COVID-19. Fecha de consulta: 25 de septiembre de 2020. Disponible en: https://www.minsalud.gov.co/Paginas/Colombia-confirma-su-primer-caso-de-COVID-19.aspx
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
World Health Organization. Laboratory biosafety guidance related to coronavirus disease (COVID-19). (COVID-19): Interim guidance, March 19, 2020. Geneva: WHO; 2020. p. 11. Fecha de consulta: 25 de septiembre de 2020. Disponible en: https://apps.who.int/iris/handle/10665/331500?locale-attribute=es&
Quick J. nCoV-2019 sequencing protocol V.1 protocols.io2020. Fecha de consulta: 25 de septiembre de 2020. Disponible en: https://www.protocols.io/view/ncov-2019-sequencingprotocol-bbmuik6w
Rambaut A, Quick J. artic-ncov2019 GitHub2020. Fecha de consulta: 25 de septiembre de 2020. Disponible en: https://github.com/artic-network/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V1/nCoV-2019_SMARTplex.tsv
Li H, Wong C, Mori Y. bwa - Burrows- Wheeler Alignment Tool 2020. Fecha de consulta: 25 de septiembre de 2020. Disponible en: http://bio-bwa.sourceforge.net/bwa.shtml
O´Toole A, McCrone J. Phylogenetic assignment of named global outbreak LINeages 2020. Fecha de consulta: 18 de junio de 2020. Disponible en: https://github.com/hCoV-2019/pangolin
Laiton-Donato K, Villabona-Arenas CJ, Usme-Ciro JA, Franco-Muñoz C, Álvarez-Díaz DA, Villabona-Arenas LS, et al. Genomic epidemiology of SARS-CoV-2 in Colombia. medRxiv. 2020;medRxiv 2020.06.26.20135715. https://doi.org/10.1101/2020.06.26.20135715
Park WB, Kwon NJ, Choi SJ, Kang CK, Choe PG, Kim JY, et al. Virus isolation from the first patient with SARS-CoV-2 in Korea. J Korean Med Sci. 2020;35:e84. https://doi.org/10.3346/jkms.2020.35.e84
Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv. 2020;bioRxiv 2020.03.02.972935. https://doi.org/10.1101/2020.03.02.972935
Centers for Disease Control and Prevention. Duration of isolation and precautions for adults with COVID-19 Healthcare workers 2020. Fecha de consulta: 25 de septiembre de 2020. Disponible en: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020;bioRxiv 2020.06.12.14872. https://doi.org/10.1101/2020.06.12.148726
Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv. 2020;bioRxiv 2020.09.01.278689. https://doi.org/10.1101/2020.09.01.278689
Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa638
Some similar items:
- Diego A. Álvarez-Díaz, Katherine Laiton-Donato, Carlos Franco-Muñoz, Marcela Mercado-Reyes, SARS-CoV-2 sequencing: The technological initiative to strengthen early warning systems for public health emergencies in Latin America and the Caribbean , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Hernando Baquero, María Elena Venegas Martinez, Lorena Velandia Forero, Fredy Neira Safi, Edgar Navarro, Neonatal late-onset infection with SARS CoV-2 , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- José Moreno-Montoya, Epidemiology of self-care beyond the individual and the sanitary spheres , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Felipe Botero-Rodríguez, Oscar Franco, Carlos, Pandemic's glossary: The ABC of coronavirus concepts , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Carolina Wiesner, Cancer research in the SARS-CoV pandemia , Biomedica: Vol. 40 No. 2 (2020)
- Alfredo G. Torres, Vaccines against SARS-CoV-2: Are they a reality for Latin America? , Biomedica: Vol. 40 No. 3 (2020)
- Zulma M. Cucunubá, Latin American scientific research prorities for COVID-19 prevention and control , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Dr., Diana Benavides-Arias, Luz Amparo Pérez, Jennifer Ruiz, Deidamia García, Iván Osejo, Edwin Ussa, Camilo Pino, Fernando Pío de La Hoz, Incidence of acute respiratory symptoms and COVID-19 infection in children in public schools in Bogotá, Colombia, from July to November, 2020 , Biomedica: Vol. 42 No. Sp. 2 (2022): Covid-19
- Juan Pablo Orozco-Hernández , Juan José Montoya-Martínez, Manuel Conrado Pacheco-Gallego, Mauricio Céspedes-Roncancio , Gloria Liliana Porras-Hurtado, SARS-CoV-2 and rhinovirus/enterovirus co-infection in a critically ill young adult patient in Colombia , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Gustavo Aroca, María Vélez-Verbel , Andrés Cadena , Lil Geraldine Avendaño , Sandra Hernández, Angélica Sierra, Omar Cabarcas, Santos Ángel Depine, COVID-19 in hemodialysis patients in Colombia: Report of seven cases , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
Funding data
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























