Clinical validation of the isothermal RT-LAMP test for rapid diagnosis of SARS-CoV-2
Abstract
Introduction: Since the first report in Wuhan (China) in 2019, the SARS-CoV-2 virus has spread throughout the world, with a significant impact in public health. To contain its transmission, the WHO has encouraged the development of rapid, simple, sensitive and specific tests that complement qRT-PCR, as the gold standard. RT-LAMP has shown to be a good alternative to detect SARS-CoV-2 in different fluid samples.
Objective: To validate the colorimetric RT-LAMP technique using two sets of primers targeting N gene of SARS-CoV-2 in 117 nasopharyngeal swab samples previously confirmed by RT-qPCR, using the Charité/Berlin protocol.
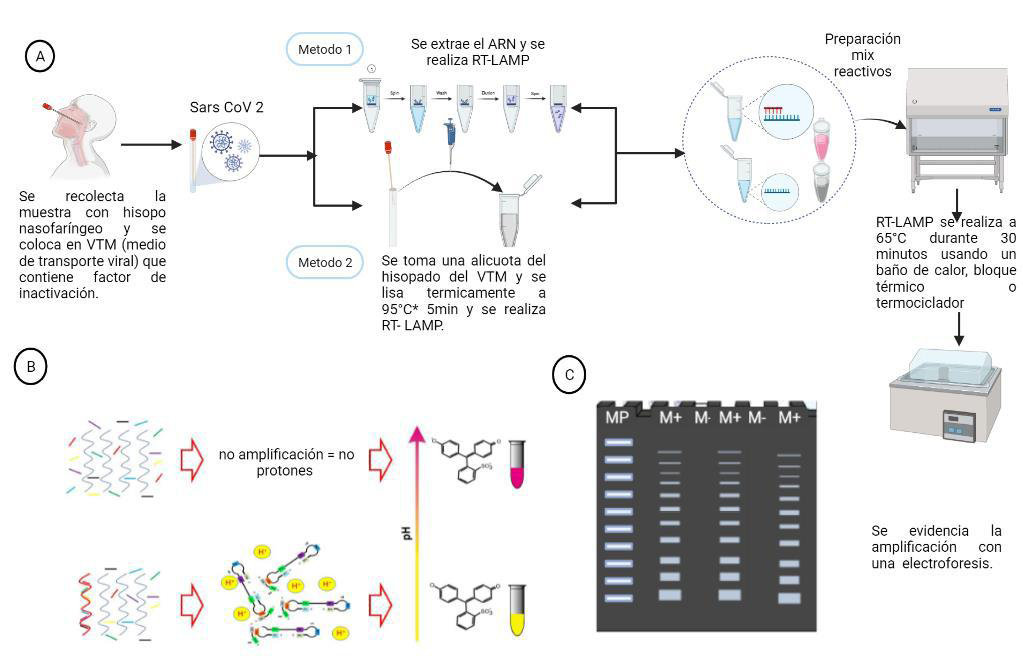
Material and methods: A total of 153 nasopharyngeal swab samples from individuals with suspected COVID-19 were subjected to qRT-PCR and RT-LAMP using a commercial
colorimetric kit (NEB, Germany). RT-LAMP was performed using both extracted RNA samples and raw samples without prior RNA extraction, and the result was assessed by a simple color change in the reaction.
Results: Sensitivity and specificity for the previously reported RT-LAMP primers (Broughton set) targeting N gene of SARS-CoV-2 were 0.97 (0.85-1.00) and 0.81 (0.65-0.92) respectively, with CI95%. The Lalli primers targeting another region of the N gene used showed a sensitivity value of 0.96 (0.78-1.00) and a specificity of 0.77 (0.55-0.92). Without RNA extraction we found a sensitivity value of 0.95 (0.74, 1.00) and a specificity of 0.88 (0.64, 0.99). A sensitivity value of 0.95 (0.74-1.00) and a specificity 0.88 (0.64-0.99) were found without prior RNA extraction.
Conclusion: Taking together, the results showed that RT-LAMP technique could be considered as a rapid diagnostic test, easy to perform, free of sophisticated equipment, sensitive and specific to diagnose SARS-CoV-2 in nasopharyngeal swabs with and without prior RNA extraction, allowing its implementation in places with scarce resources.
Downloads
References
Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533-4. https://doi.org/10.1016/S1473-3099(20)30120-1
Chiem K, Morales-Vásquez D, Park JG, Platt RN, Anderson T, Walter MR, et al. Generation and characterization of recombinant SARS-CoV-2 expressing reporter genes. J Virol. 2021;95:e02209-20. https://doi.org/10.1128/JVI.02209-20
Jegerlehner S, Suter-Riniker F, Jent P, Bittel P, Nagler M. Diagnostic accuracy of a SARSCoV-2 rapid antigen test in real-life clinical settings. Int J Infect Dis. 2021;109:118-22. https://doi.org/10.1016/j.ijid.2021.07.010
Maricic T, Nickel O, Aximu-Petri A, Essel E, Gansauge M, Kanis P, et al. A direct RTqPCR approach to test large numbers of individuals for SARS-CoV-2. PLoS ONE. 2020;15:e0244824. https://doi.org/10.1371/journal.pone.0244824
Mustafa MI, Makhawi AM. SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases. J Clin Microbiol. 2021;59:e00745-20. https://doi.org/10.1128/JCM.00745-20
Xiong D, Dai W, Gong J, Li G, Liu N, Wu W, et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLOS Biol. 2020;18:e3000978. https://doi.org/10.1371/journal.pbio.3000978
Ghosh P, Chowdhury R, Hossain ME, Hossain F, Miah M, Rashid MdU, et al. Evaluation of recombinase-based isothermal amplification assays for point-of-need detection of SARSCoV-2 in resource-limited settings. Int J Infect Dis.;114:105-11. https://doi.org/10.1016/j.ijid.2021.11.007
Liang Y, Lin H, Zou L, Zhao J, Li B, Wang H, et al. CRISPR-Cas12a-based detection for the major SARS-CoV-2 variants of concern. Microbiol Spectr. 2021;9:e01017-21. https://doi.org/10.1128/Spectrum.01017-21
Ma L, Yin L, Li X, Chen S, Peng L, Liu G, et al. A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosens Bioelectron. 2022;195:113646. https://doi.org/10.1016/j.bios.2021.113646
Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223-9. https://doi.org/10.1006/mcpr.2002.0415
Tanner NA, Zhang Y, Evans TC. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques. 2015;58:59-68. https://doi.org/10.2144/000114253
Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcriptionloop-mediated isothermal amplification. PLoS ONE. 2020;15:e0234682. https://doi.org/10.1371/journal.pone.0234682
Fischbach J, Xander NC, Frohme M, Glökler JF. Shining a light on LAMP assays. A comparison of LAMP visualization methods including the novel use of berberine. BioTechniques. 2015;58:189-94. https://doi.org/10.2144/000114275
Park GS, Ku K, Baek SH, Kim SJ, Kim SI, Kim BT, et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J Mol Diagn. 2020;22:729-35. https://doi.org/10.1016/j.jmoldx.2020.03.006
Notomi T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63e-63. https://doi.org/10.1093/nar/28.12.e63
Aliotta JM, Pelletier JJ, Ware JL, Moran LS, Benner JS, Kong H. Thermostable Bst DNA polymerase I lacks a 3’-->5’ proofreading exonuclease activity. Genet Anal Biomol Eng. 1996;12:185-95. https://doi.org/10.1016/S1050-3862(96)80005-2
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870-4. https://doi.org/10.1038/s41587-020-0513-4
Lalli MA, Langmade JS, Chen X, Fronick CC, Sawyer CS, Burcea LC, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem. 2021;67:415-24. https://doi.org/10.1093/clinchem/hvaa267
Aranha C, Patel V, Bhor V, Gogoi D. Cycle threshold values in RT-PCR to determine dynamics of SARS-CoV-2 viral load: An approach to reduce the isolation period for COVID-19 patients. J Med Virol. 2021;93:6794-7. https://doi.org/10.1002/jmv.27206
Soni S, Salhotra A, Suar M. Handbook of research on diverse applications of nanotechnology in biomedicine, chemistry, and engineering. Hershey, PA: Engineering Science Reference; 2015. p. 820.
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159-74. https://doi.org/10.2307/2529310
Wei S, Kohl E, Djandji A, Morgan S, Whittier S, Mansukhani M, et al. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. Sci Rep. 2021;11:2402. https://doi.org/10.1038/s41598-021-81487-y
Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, et al. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62:41-8. https://doi.org/10.1111/j.1574-695X.2011.00785.x
Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRciv. 2020. https://doi.org/10.1101/2020.02.26.20028373
Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, et al. A colorimetric RTLAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12:eabc7075. https://doi.org/10.1126/scitranslmed.abc7075
Flynn MJ, Snitser O, Flynn J, Green S, Yelin I, Szwarcwort-Cohen M, et al. A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRciv. 2020. https://doi.org/10.1101/2020.11.19.20234948
Savela ES, Viloria Winnett A, Romano AE, Porter MK, Shelby N, Akana R, et al. Quantitative SARS-CoV-2 viral-load curves in paired saliva samples and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J Clin Microbiol. 2022;60:e01785-21. https://doi.org/10.1128/jcm.01785-21
Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12:863. https://doi.org/10.3390/v12080863
Some similar items:
- César Antonio Bonilla-Asalde, David Alberto Díaz-Robles, Edwin César Cieza-Macedo, Oriana Rivera-Lozada, Lisandro A. Pacheco, Leidy Hurtado, Diana Díaz, Katherine Escorcia, Laura Flórez, Yesit Bello, Yirys Díaz, Elkin Navarro, Leonardo C. Pacheco, Nataly Galán, Ronald Maestre, Antonio Acosta, Letter to the editor. Clinical validation of the isothermal RT-LAMP test for rapid diagnosis of SARS-CoV-2. Biomédica. 2022;42(Supl.2):59-72. , Biomedica: Vol. 43 No. 1 (2023)
- Astrid Elena Montoya, José Menco, Natalia Osorio, Maria Alejandra Zuluaga, Juliana Duque, Giovanny Torres, Marcos Restrepo, Concordance between thick blood smear, immunochromatography and polymerase chain reaction for malaria diagnosis , Biomedica: Vol. 28 No. 2 (2008)
- Ruth Aralí Martínez-Vega, Fredi Alexander Díaz Quijano, Carolina Coronel Ruiz, Sergio Yebrail Gómez, Luis Ángel Villar Centeno, Evaluation of PANBIO rapid immunochromatographic cassette for dengue diagnosis in a Colombian endemic area , Biomedica: Vol. 29 No. 4 (2009)
- Nohora Marcela Mendoza, Marisol García, Liliana Jazmín Cortés, Claudia Vela, Rigoberto Erazo, Pilar Pérez, Olga Lucía Ospina, Javier Darío Burgos, Evaluation of two rapid diagnostic tests, NOW® ICT Malaria Pf/Pv and OptiMAL®, for diagnosis of malaria , Biomedica: Vol. 27 No. 4 (2007)
- Álvaro Sanabria, Luis Carlos Domínguez, Charles Bermúdez, Adriana Serna, Evaluation of diagnostic scales for appendicitis in patients with lower abdominal pain , Biomedica: Vol. 27 No. 3 (2007)
- Adriana Pabón, Gonzalo Álvarez, Jorge Yánez, Carlos Céspedes, Yensa Rodríguez, Ángela Restrepo, Silvia Blair, Evaluation of ICT malaria immunochromatographic Binax NOW® ICT P.f/P.v test for rapid diagnosis of malaria in a Colombian endemic area , Biomedica: Vol. 27 No. 2 (2007)
- María Victoria Benjumea, Diagnostic accuracy of five gestational references to predict insufficient birth weight , Biomedica: Vol. 27 No. 1 (2007)
- Nelson José Alvis-Zakzuk, María de los Ángeles Carrasquilla, Verónica Jhajaira Gómez, Jaime Robledo, Nelson Rafael Alvis-Guzmán, José Mauricio Hernández, Diagnostic accuracy of three technologies for the diagnosis of multi-drug resistant tuberculosis , Biomedica: Vol. 37 No. 3 (2017)
- Lina Ruiz, María Angélica Maya, Zulma Vanesa Rueda, Lucelly López, Lázaro Agustín Vélez, Current characteristics of tuberculosis and human immunodeficiency virus co-infection in a cohort of hospitalized patients in Medellín, Colombia , Biomedica: Vol. 38 No. Sup. 2 (2018): Suplemento 2, Medicina tropical
- Freddy Agredo , Lyda Osorio , Coverage and fidelity of the Xpert MTB/RIF™ implementation in a high-burden area for pulmonary tuberculosis in Colombia , Biomedica: Vol. 40 No. 4 (2020)
Copyright (c) 2022 Biomedica

This work is licensed under a Creative Commons Attribution 4.0 International License.
Funding data
-
Sistema General de Regalías de Colombia
Grant numbers BPIN #2020000100144 -
Departamento Administrativo de Ciencia, Tecnología e Innovación (COLCIENCIAS)
Grant numbers contrato #462
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























