Genomic epidemiology of SARS-CoV-2 δ sublineages of the second wave of 2021 in Antioquia, Colombia
Abstract
Introduction. During the development of the SARS-CoV-2 pandemic in Antioquia, we experienced epidemiological peaks related to the α, ɣ, β, ƛ, and δ variants. δ had
the highest incidence and prevalence. This lineage is of concern due to its clinical manifestations and epidemiological characteristics. A total of 253 δ sublineages have been reported in the PANGOLIN database. The sublineage identification through genomic analysis has made it possible to trace their evolution and propagation.
Objective. To characterize the genetic diversity of the different SARS-CoV-2 δ sublineages in Antioquia and to describe its prevalence.
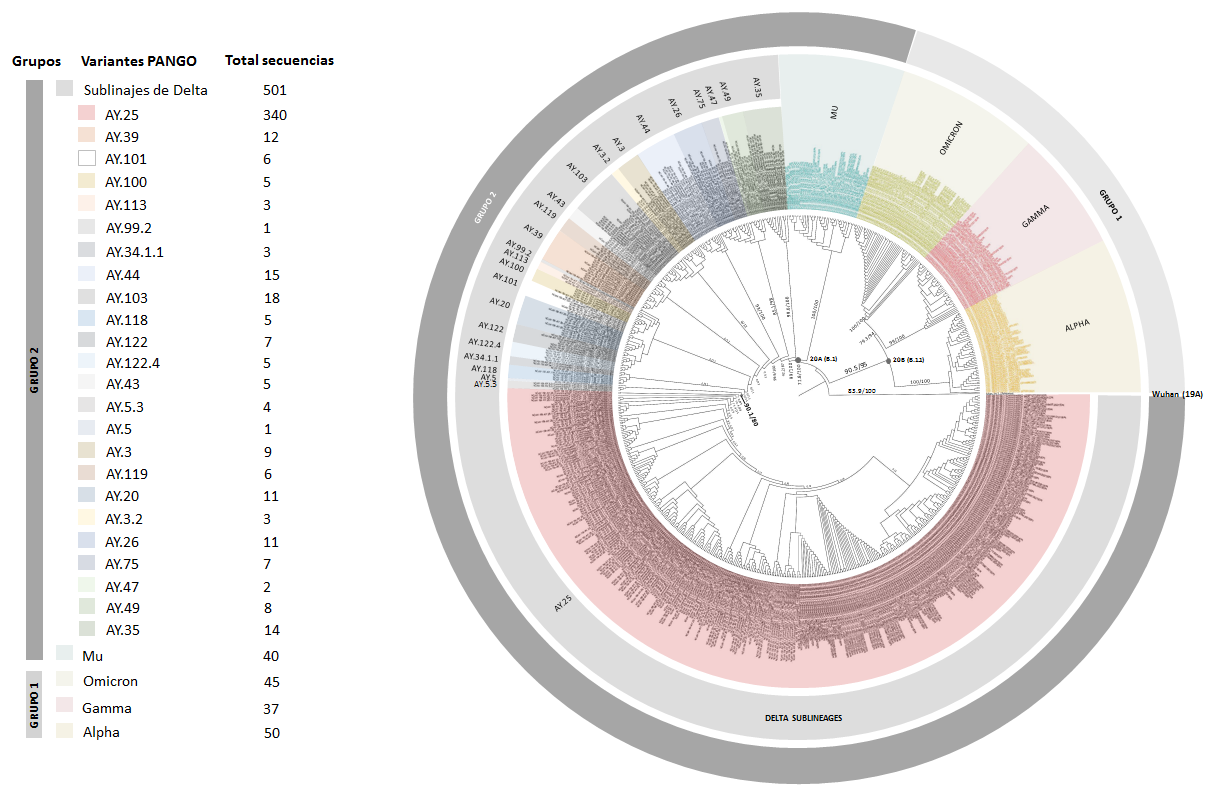
Materials and methods. We collected sociodemographic information from 2,675 samples, and obtained 1,115 genomes from the GISAID database between July 12th, 2021, and January 18th, 2022. From the analyzed genomes, 515 were selected because of their high coverage values (>90%) to perform phylogenetic analysis and to infer allele frequencies of mutations of interest.
Results. We characterized 24 sublineages. The most prevalent was AY.25. Mutations of interest as L452R, P681R, and P681H were identified in this sublineage, comprising a frequency close to 0.99.
Conclusions. This study identified that the AY.25 sublineage has a transmission advantage compared to the other δ sublineages. This attribute may be related to the presence of the L452R and P681R mutations associated in other studies with higher evasion of the immune system and less efficacy of drugs against SARS-CoV-2.
Downloads
References
Instituto Nacional de Salud. Casos COVID 19. Colombia, 2020 - 2022. Fecha de consulta: 25 de abril de 2022. Disponible en: https://app.powerbi.com/viewr=eyJrIjoiMjBjZWNlOGUtNzc1Yi00NjVkLTkyMjktOTJmMGU3YTU2Nzk4IiwidCI6ImE2MmQ2YzdiLTlmNTktNDQ2OS05MzU5LTM1MzcxNDc1OTRiYiIsImMiOjR9&pageName=ReportSection0c50ea3406afe4407370
Laiton-Donato K, Villabona-Arenas CJ, Usme-Ciro JA, Franco-Muñoz C, Álvarez-Díaz DA, Villabona-Arenas LS, et al. Genomic epidemiology of severe acute respiratory syndrome coronavirus 2, Colombia. Emerg Infect Dis. 2020;26: 2854-62. https://doi.org/10.3201/eid2612.202969
Velásquez Cuadros CH, Almanza Payares RE, Silva López YJ. Clinical, epidemiological and virological findings of the first COVID-19 outbreak in Medellín, Antioquia, Colombia. Rev Fac Nac Salud Pública. 2022;40:e344675. https://doi.org/10.17533/udea.rfnsp.e344675
Betancur I, Velarde CA, Ortiz C. Situación epidemiológica de las variantes de SARSCoV-2 detectadas en Antioquia desde diciembre 2020 a enero 2022. Fecha de consulta: 25 de abril de 2022. Disponible en: https://www.dssa.gov.co/images/2022/documentos/investigaciones/Epidemiologia_de_SARS_CoV-2_en_Antioquia_2020-2022.pdf
Laboratoire National de Référence pour les Infections Respiratoires Aiguës. Delta variant B.1.617.2 lineage and AY sublineages. 2021. Fecha de consulta: 19 de octubre de 2022. Disponible en: https://lns.lu/wp-content/uploads/2021/09/lns-scientific-briefing-deltasublineages_final_en_v20210913.pdf
World Health Organization. COVID-19 weekly epidemiological update, 31 August 2021.2021. Fecha de consulta: 19 de octubre de 2022. Disponible en: https://apps.who.int/iris/handle/10665/344799?locale-attribute=es&
Taboada B, Zárate S, García-López R, Muñoz-Medina JE, Sánchez-Flores A, Herrera-Estrella A, et al. Dominance of three sublineages of the SARSCoV-2 delta variant in Mexico. Viruses. 2022;14:1165. https://doi.org/10.3390/v14061165
Elliott P, Haw D, Wang H, Eales O, Walters CE, Ainslie KEC, et al. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the delta variant. Science. 2021;374:eabl9551. https://doi.org/10.1126/science.abl9551
Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARSCoV2 spike mutations, l452r, t478k, e484q and p681r, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. https://doi.org/10.3390/microorganisms9071542
Instituto Nacional de Salud. Caracterización genómica de SARS-CoV-2 por muestreo probabilístico en Colombia - Tercer muestreo periodo octubre de 2021. Bogotá: Instituto Nacional de Salud; 2021. Disponible en: http://www.ins.gov.co/BibliotecaDigital/Tercer_muestreo-Estrategia-caracterizacion-genomica-muestreo-probabilistico-SARS-CoV-2.pdf
Betancur I, Velarde CA, Ortiz C. Situación epidemiológica de las variantes de Sars-CoV-2 detectadas en Antioquia desde diciembre 2020 a enero 2022. Boletín Epidemiológico de Antioquia. 2021. Disponible en: https://www.dssa.gov.co/images/2022/documentos/investigaciones/Epidemiologia_de_SARS_CoV-2_en_Antioquia_2020-2022.pdf
Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403. https://doi.org/10.1038/s41564-020-0770-5
Garcia-Beltran WF, St. Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARSCoV-2 ómicron variant. Cell. 2022;185:457. https://doi.org/10.1101/2021.12.14.21267755
Modes ME, Directo MP, Melgar M, Johnson LR, Yang H, Chaudhary P, et al. Clinical characteristics and outcomes among adults hospitalized with laboratory-confirmed SARSCoV-2 infection during periods of B.1.617.2 (delta) and B.1.1.529 (ómicron) variant predominance — One Hospital, California, July 15-September 23, 2021, and December 21, 2021–January 27, 2022. MMWR Recomm Reports. 2022;71:217-23. https://doi.org/10.15585/mmwr.mm7106e2
Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data – From vision to reality. Euro Surveill. 2017;22:30494. https://doi.org/10.2807/1560-7917.ES.2017.22.13.30494
Artic Networ. Artic Network. 2019. Fecha de consulta: 11 de agosto de 2022. Disponible en: https://artic.network/ncov-2019/ncov2019-it-setup.html
Quick J. nCoV-2019 sequencing protocol V.1. Protocolo.io. 2021. Fecha de consulta: 19 de octubre de 2022. Disponible en: https://www.protocols.io/view/ncov-2019-sequencingprotocol-bp2l6n26rgqe/v1?version_warning=no
Márquez S, Prado-Vivar B, Guadalupe JJ, Gutiérrez-Granja B, Jibaja M, Tobar M, et al. Genome sequencing of the first SARS-CoV-2 reported from patients with COVID-19 in Ecuador. medRxiv. 2020:2020.06.11.20128330. https://doi.org/10.1101/2020.06.11.20128330
IBM. SPSS Statistics. Fecha de consulta: 19 de octubre de 2022. Disponible en: https://www.ibm.com/mysupport/s/topic/0TO500000001yjtGAA/spss-statistics?language=en_US.2022 [citado 19 de octubre de 2022]. Disponible en: https://www.ibm.com/mysupport/s/topic/0TO500000001yjtGAA/spss-statistics?language=en_US
Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:5:113. https://doi.org/10.1186/1471-2105-5-113
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972-3. https://doi.org/10.1093/bioinformatics/btp348
Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis, version 11. Mol Biol Evol. 2021;38 :3022-7. https://doi.org/10.1093/molbev/msab120
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530-4. https://doi.org/10.1093/molbev/msaa015
Sielemann K, Pucker B, Schmidt N, Viehöver P, Weisshaar B, Heitkam T, et al. Complete pan-plastome sequences enable high resolution phylogenetic classification of sugar beet and closely related crop wild relatives. BMC Genomics. 2022;23:113. https://doi.org/10.1186/s12864-022-08336-8
Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422-3. https://doi.org/10.1093/bioinformatics/btp163
Jiménez-Silva C, Rivero R, Douglas J, Bouckaert R, Villabona-Arenas CJ, Atkins KE, et al. Genomic epidemiology of SARS-CoV-2 variants during the first two years of the pandemic in Colombia. Commun Med (Lond). 2023;3:97. https://doi.org/10.1038/s43856-023-00328-3
Murall CL, Poujol R, Petkau A, Sobkowiak B, Zetner A, Kraemer SA, et al. Monitoring the evolution and spread of delta sublineages AY.25 and AY.27 in Canada. 2021. Fecha de consulta: 19 de octubre de 2022. Disponible en: https://virological.org/t/monitoring-theevolution-and-spread-of-delta-sublineages-ay-25-and-ay-27-in-canada/767
Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, et al. Outbreak of SARSCoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings — Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1059-62. https://doi.org/10.15585/mmwr.mm7031e2
Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. NextStrain: Realtime tracking of pathogen evolution. Bioinformatics. 2018;34:4121-3. https://doi.org/10.1093/bioinformatics/bty407
Rabi FA, Al Zoubi MS, Al-Nasser AD, Kasasbeh GA, Salameh DM. SARSCoV-2 and coronavirus disease 2019: What we know so far. Pathogens. 2020;9:231. https://doi.org/10.3390/pathogens9030231
Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124-36. https://doi.org/10.1016/j.chom.2021.06.006
Selzer T, Albeck S, Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat Struct Biol. 2000;7:537-41. https://doi.org/10.1038/76744
Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021:2021.03.07.21252647. https://doi.org/10.1101/2021.03.07.21252647
Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over alfa variant. Cell Rep. 2022;39:110829. https://doi.org/10.1016/j.celrep.2022.110829
Pastrian-Soto G, Pastrian-Soto G. Presencia y expresión del receptor ACE2 (Target de SARS-CoV-2) en tejidos humanos y cavidad oral. Posibles rutas de infección en órganos orales. Int J Odontostomatol. 2020;14:501-7. https://doi.org/10.4067/S0718-381X2020000400501
Lubinski B, Fernandes MH V., Frazier L, Tang T, Daniel S, Diel DG, et al. Functional evaluation of the P681H mutation on the proteolytic activation the SARS-CoV-2 variant B.1.1.7 (Alfa) spike. iScience. 2022;25:103589. https://doi.org/10.1016/j.isci.2021.103589
Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, et al. SARSCoV2 neutralizing antibody LY-CoV555 in outpatients with COVID19. N Engl J Med. 2021;384:229-37. https://doi.org/10.1056/NEJMoa2029849
Liu H, Wei P, Kappler JW, Marrack P, Zhang G. SARS-CoV-2 Variants of concern and variants of interest receptor binding domain mutations and virus infectivity. Front Immunol. 2022;13:825256. https://doi.org/10.3389/fimmu.2022.825256
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44. https://doi.org/10.1016/j.chom.2020.11.007
Some similar items:
- Daniel Eduardo Henao, Fabián Alberto Jaimes, Evidence-based medicine: an epistemological approach , Biomedica: Vol. 29 No. 1 (2009)
- Amanda Maestre, Jaime Carmona-Fonseca, Amanda Maestre, Alta frecuencia de mutaciones puntuales en pfcrt de Plasmodium falciparum y emergencia de nuevos haplotipos mutantes en Colombia , Biomedica: Vol. 28 No. 4 (2008)
- Luz Elena Velásquez, Catalina Gómez, Erika Valencia, Laura Salazar, Eudoro Casas, Paragonimosis in the peri-urban zone of Medellín, Antioquia , Biomedica: Vol. 28 No. 3 (2008)
- John Alexander Galindo, Fabio Aníbal Cristiano, Angélica Knudson, Rubén Santiago Nicholls, Ángela Patricia Guerra, Point mutations in dihydrofolate reductase and dihydropteroate synthase genes of Plasmodium falciparum from three endemic malaria regions in Colombia , Biomedica: Vol. 30 No. 1 (2010)
- Juan P. Gómez, Juan C. Quintana, Patricia Arbeláez, Jorge Fernández, Juan F. Silva, Jacqueline Barona, Juan C. Gutiérrez, Abel Díaz, Rafael Otero, Tityus asthenes scorpion stings: epidemiological, clinical and toxicological aspects , Biomedica: Vol. 30 No. 1 (2010)
- Juan Gabriel Piñeros, Malaria and social health determinants: a new heuristic framework from the perspective of Latin American social medicine , Biomedica: Vol. 30 No. 2 (2010)
- Richard Hoyos, Lisandro Pacheco, Luz Adriana Agudelo, German Zafra, Pedro Blanco, Omar Triana, Seroprevalence of Chagas disease and associated risk factors in a population of Morroa, Sucre , Biomedica: Vol. 27 No. 1esp (2007): Enfermedad de Chagas
- Diana Marcela Echeverry, Juan Miguel Rengifo, Jhon Carlos Castaño, Germán Alberto Téllez, María Mercedes González, Prevalence of Mammomonogamus laryngeus (Strongylida: Syngamidae) nematodes in a bovine slaughterhouse , Biomedica: Vol. 31 No. 3 (2011)
- Gladys Acuña-González, Carlo E. Medina-Solís, Gerardo Maupomé, Mauricio Escoffie-Ramírez, Jesús Hernández-Romano, María de L. Márquez-Corona, Arturo J. Islas-Márquez, Juan J. Villalobos-Rodelo, Family history and socioeconomic risk factors for non-syndromic cleft lip and palate: A matched case-control study in a less developed country , Biomedica: Vol. 31 No. 3 (2011)
- Pablo Chaparro, Julio Padilla, Malaria mortality in Colombia, 1979-2008 , Biomedica: Vol. 32 (2012): Suplemento 1, Malaria
Copyright (c) 2024 Biomedica

This work is licensed under a Creative Commons Attribution 4.0 International License.
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























