Methicillin resistance and biofilm production in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus in México
Abstract
Introduction: Infections associated with health care caused by S. aureus and coagulase-negative Staphylococci multi-resistant to antibiotics cause a high epidemiological impact due to their high morbidity and mortality. Biofilm formation, which has been associated with antimicrobial resistance, can also occur.
Objectives: To determine methicillin resistance and to quantify the biofilm production to establish if there is a relationship in clinical isolates of S. aureus and coagulase-negative Staphylococci.
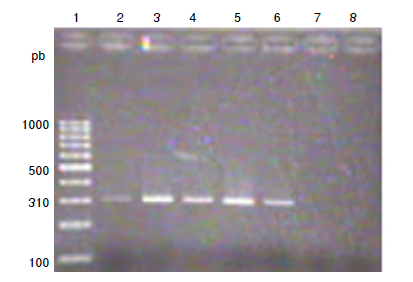
Material and methods: A total of 11 strains of S. aureus and 12 of coagulase-negative Staphylococci were studied. Methicillin resistance was determined with cefoxitin discs and the Clinical Laboratory Standards Institute (CSLI), 2018 reference values. Biofilm production was quantified by the crystal violet method. The mecA and icaADBC genes were identified by PCR. A bivariate analysis was performed with chi-square (c2) and Cramér’s V statistical tests, using SPSS™, version 20.0 software.
Results: Nine S. aureus strains were methicillin-resistant and two were sensitive. Eight coagulase-negative Staphylococci strains were resistant and four were sensitive. The mecA genotype was found in eight of the nine S. aureus resistant strains and six of eight resistant coagulase-negative Staphylococci. All strains formed biofilms. Ten strains of S. aureus and 11 of coagulase-negative Staphylococci presented the icaADCB genotype. No association was found between methicillin-resistance and biofilm formation.
Conclusions: Cefoxitin is enough to define the resistance phenotype and is associated with the mecA genotype. All strains formed biofilms and were related to the presence of the icaADCB operon. Biofilm formation and methicillin resistance were independent features in both groups of strains.
Downloads
References
Guzmán-Blanco M, Mejía C, Isturiz R, Álvarez C, Bavestrello L, Gotuzzo E, et al. Epidemiology of meticillin resistant Staphylococcus aureus (MRSA) in Latin America. Int J Antimicrob Agents. 2009;34:304-8. https://doi.org/10.1016/j.ijantimicag.2009.06.005
Red Hospitalaria de Vigilancia Epidemiológica. Informe anual 2015. México: Secretaría de Salud; 2015.
Bannerman TL, Peacock SJ. Staphylococcus, Micrococcus, and other catalase-positive cocci. En: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of clinical microbiology. 9th edition. Washington, D.C.: ASM Press; 2007. p. 390-411.
Bustos-Martínez JA, Hamdan-Partida A, Gutiérrez-Cárdenas. Staphylococcus aureus: la reemergencia de un patógeno en la comunidad. Rev Biomed. 2006;17:287-305.
Velázquez-Meza ME. Surgimiento y diseminación de Staphylococcus aureus meticilino resistente. Salud Pública de México. 2005;47:381-7.
Marín M, Gudiol F. Antibióticos beta-lactámicos. Enferm Infecc Microbiol Clin. 2003;21:42-55. https://doi.org/10.1016/S0213-005X(03)72873-0
Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629-41. https://doi.org/10.1038/nrmicro
Zhang HZ, Hackbart CJ, Chansky KM, Chambers HF. A proteolytic transmembrane pathway and resistance to β lactams in staphylococci. Science. 2001;291:1962-5. https://doi.org/10.1126/science.1055144
Hiramatsu K. Vancomycin resistant Staphylococcus aureus: A new model of antibiotic resistance. Lancet Infec Dis. 2001;1:147-55 https://doi.org/10.1016/S1473-3099(01)00091-3
Hernández JB, Novales GM. Biofilm. ¿Amenaza latente o factor de protección? Estado de arte. Enferm Infecc Microbiol Clin. 2007;27:22-8.
Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95-108. https://doi.org/10.1038/nrmicro821
Otto M. Staphylococcus epidermidis the “accidental” pathogen. Nat Rev Microbiol. 2009;7:555-67. https://doi.org/10.1038/nrmicro2182
Lasa I. Towards the identification of the common features of bacterial biofilm development. Int Microbiol. 2006;9:21-8.
O´Gara PJ. ica and beyond: Biofilms mechanism and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179-88. https://doi.org/10.1111/j.1574-6968.2007.00688.x
Cucarella C, Tormo MA, Knecht E, Amorena B, Lasa I, Foster TJ, et al. Expression of the biofilm associate protein interferes with host protein receptors of Staphylococcus aureus and alters the infective process. Infect Immun. 2002;70:3180-8. https://doi.org/10.1128/IAI.70.6.3180-3186.2002
Rodríguez-Martínez JM, Pascual Á. Actividad de los antimicrobianos en biocapas bacterianas. Enferm Infecc Microbiol Clin. 2008;26:107-14. https://doi.org/10.1157/13115546
Clinical and Laboratory Standards Institute. Antimicrobial susceptibility testing standards. M02-A11, M07-A9, and M11-A8. Wayne: ILSI; 2014.
Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40:175-9. https://doi.org/10.1016/S0167-7012(00)00122-6
García-Barreto AA. Biofilm de Staphylococcus spp. de origen intrahospitalario: genes asociados (tesis). México: Instituto Politécnico Nacional; 2010.
Acosta-Pérez G, Rodríguez-Ábrego G, Longoria-Revilla E, Castro-Mussot ME. Evaluación de cuatro métodos para la detección de Staphylococcus aureus meticilino-resistente de muestras clínicas en un hospital regional. Salud Pública de México. 2012;54:1-6.
Gómez J, Gómez-Lus ML, Bas P, Ramos C, Cafini F, Maestre JR, et al. ¿Es la cuantificación del biofilm un elemento diferenciador en la patogenia de bacilos gramnegativos? Rev Esp Quimioter. 2013;26:97-102.
Ducel G, Fabry J, Nicolle L. Prevención de las infecciones nosocomiales. Guía práctica. Segunda edición. México: Organización Mundial de la Salud; 2003. p. 1-65.
Zayas-Tamayo ÁM, Barreras-García G, Álvarez-Varela E. Detección mediante el Sistema DIRAMIC de Staphylococcus aureus resistente a meticilina (SARM) y comparación con otros métodos utilizado en la práctica clínica. Revista CENIC Ciencias Biológicas. 2013;44. Fecha de consulta: 23 de noviembre de 2017. Disponible en: https://revista.cnic.edu.cu/revistaCB/sites/default/files/articulos/CB28-12.pdf
Corso A, Soloaga R, Faccone D, Gagetti P, Corbella S, Iglesias M, et al. Improvement of a latex agglutination test for the evaluation of oxacillin resistance in coagulase-negative staphylococci. Diagn Microbiol Infect Dis. 2004;50:223-5. https://doi.org/10.1016/j.diagmicrobio.2004.06.005
Peeters E, Nelis HJ, Coenye T. Comparison of multiplex methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2007;11:9-18. https://doi.org/10.1016/j.mimet.2007.11.010
Arslan S, Ozkardes F. Slime production and antibiotic susceptibility in Staphylococci isolated from clinical samples. Mem Inst Oswaldo Cruz. 2007;102:29-33. https://doi.org/10.1590/S0074-02762007000100004
Smith K, Pérez A, Ramage G, Lappin D, Gemmell CG, Lang S. Biofilm formation by Scottish clinical isolates of Staphylococcus aureus. J Med Microb. 2008;57:1018-23. https://doi.org/10.1099/jmm.0.2008/000968-0
Some similar items:
- José Alejandro Martínez-Ibarra, Jorge Alejandro Martínez-Grant, Miguel Roberto Verdugo-Cervantes, Rafael Bustos-Saldaña, Benjamín Nogueda-Torres, Monitoring triatomid bug (Hemiptera: Reduviidae) presence by sentinel chicken coops in Southern Jalisco State, México , Biomedica: Vol. 30 No. 1 (2010)
- Narda María Olarte, Ismael Alberto Valderrama, Karlo Roberto Reyes, Martha Isabel Garzón, Javier Antonio Escobar, Betsy Esperanza Castro, Natasha Vanegas, Methicillin-resistant Staphylococcus aureus colonization in a Colombian hospital intensive care unit: phenotypic and molecular characterization , Biomedica: Vol. 30 No. 3 (2010)
- Ana Victoria Suescún, Juan Rodrigo Cubillos, María Mercedes Zambrano, Genes involved in fimbrial biogenesis affect biofilm formation in Klebsiella pneumoniae , Biomedica: Vol. 26 No. 4 (2006)
- Gladys Acuña-González, Carlo E. Medina-Solís, Gerardo Maupomé, Mauricio Escoffie-Ramírez, Jesús Hernández-Romano, María de L. Márquez-Corona, Arturo J. Islas-Márquez, Juan J. Villalobos-Rodelo, Family history and socioeconomic risk factors for non-syndromic cleft lip and palate: A matched case-control study in a less developed country , Biomedica: Vol. 31 No. 3 (2011)
- Javier Antonio Escobar, Ingrid Tatiana Gómez, Martha Johanna Murillo, Betsy Esperanza Castro, Bibiana Chavarro, Ricaurte Alejandro Márquez, Natasha Vanegas, Design of two molecular methodologies for the rapid identification of Colombian community-acquired methicillin-resistant Staphylococcus aureus isolates , Biomedica: Vol. 32 No. 2 (2012)
- Juan José Villalobos Rodelo, Carlo Eduardo Medina Solís, Nelly Molina Frechero, Ana Alicia Vallejos Sánchez, América Patricia Pontigo Loyola, José Luis Espinoza Beltrán, Dental caries in schoolchildren aged 6-12 years in Navolato, Sinaloa, México: experience, prevalence, severity and treatment needs. , Biomedica: Vol. 26 No. 2 (2006)
- Mayra Alejandra Machuca, Clara Isabel González, Luis Miguel Sosa, Methicillin-resistant Staphylococcus aureus causes both community-associated and health care-associated infections in children at the Hospital Universitario de Santander , Biomedica: Vol. 34 (2014): Abril, Suplemento 1, Resistencia bacteriana
- Natalia Andrea Maldonado, María Isabel Múnera, Jaime Alberto López, Patricia Sierra, Carlos Robledo, Jaime Robledo, Grupo GERMEN, Trends in antibiotic resistance in Medellín and municipalities of the Metropolitan Area between 2007 and 2012: Results of six years of surveillance , Biomedica: Vol. 34 No. 3 (2014)
- Javier Antonio Escobar-Pérez, Betsy Esperanza Castro, Ricaurte Alejandro Márquez-Ortiz, Sebastián Gaines, Bibiana Chavarro, Jaime Moreno, Aura Lucía Leal, Natasha Vanegas, Methicillin-sensitive Staphylococcus aureus isolates related to USA300 clone: Origin of community-genotype MRSA in Colombia? , Biomedica: Vol. 34 (2014): Abril, Suplemento 1, Resistencia bacteriana
- Liliana I. Barrero, Juan S. Castillo, Aura L. Leal, Ricardo Sánchez, Jorge A. Cortés, Carlos A. Álvarez, Andrés L. González, Economic burden of methicillin-resistant Staphylococcus aureus bacteremia in critical care patients in hospitals in Bogotá , Biomedica: Vol. 34 No. 3 (2014)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























