Phylogeny and antimicrobial resistance of extended-spectrum beta-lactamaseproducing Escherichia coli from hospitalized oncology patients in Perú
Abstract
Introduction: Healthcare-associated infections are a public health problem due to the increased morbimortality of patients, especially those with risk factors such as immunosuppression due to oncological diseases. It is essential to determine the genetic diversity of the main microorganisms causing healthcare infections by combining traditional epidemiological surveillance and molecular epidemiology for better outbreak follow-up and early detection.
Objective: To determine the phylogenetic group and antibiotic resistance of Escherichia coli isolated from hospitalized oncologic patients.
Materials and methods: We conducted a cross-sectional study of 67 strains of ESBL-producing Escherichia coli to determine their phylogenetic group and described their antibiotic resistance profile, beta-lactam resistance genes, as well as the type of sample and the hospitalization areas from which they were recovered.
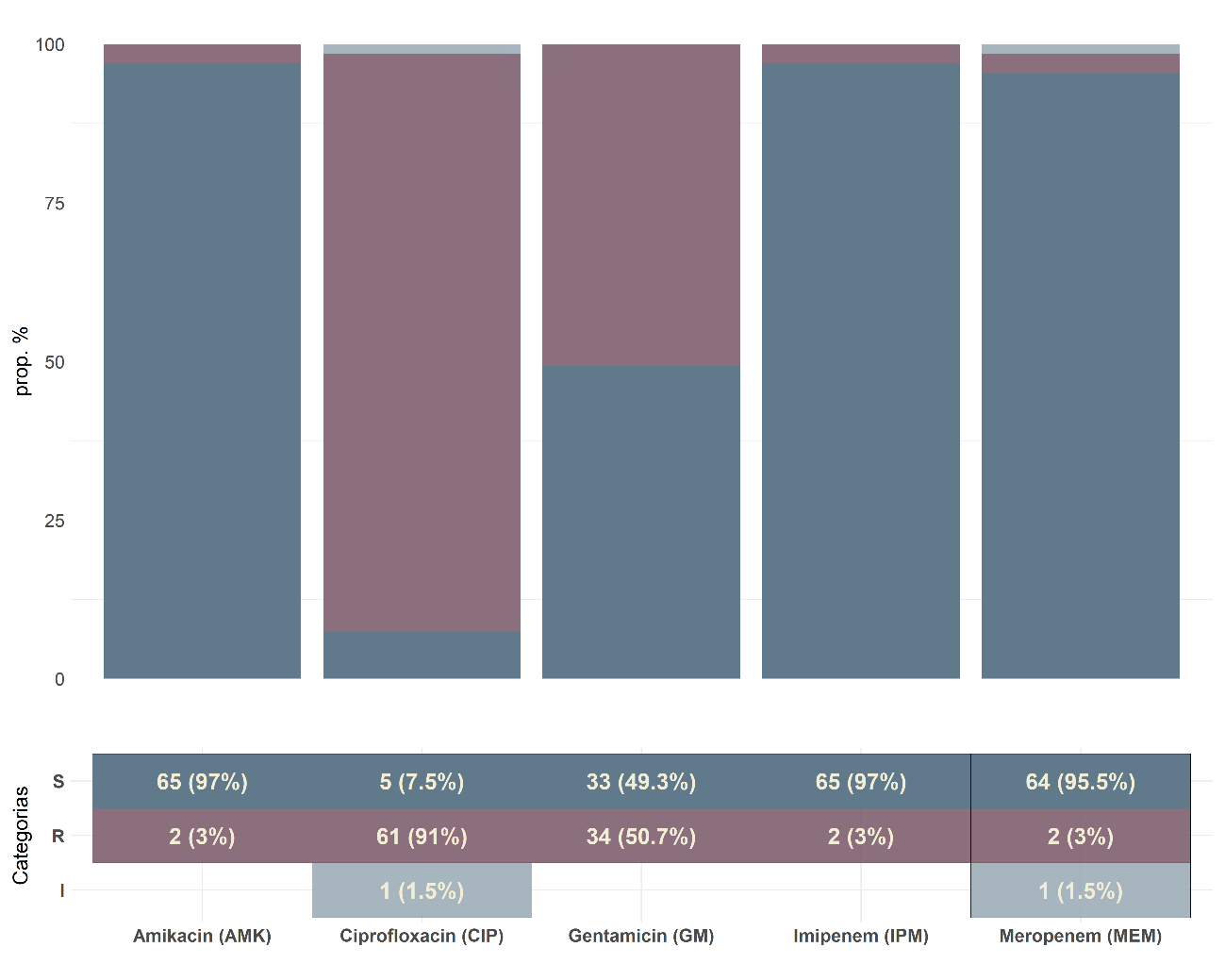
Results: The most frequent phylogenetic group was B2 (36%); 57% of B2 strains were isolated from urine and 33% came from the urology department. Resistance to ciprofloxacin and gentamicin was 92% and 53%, respectively, and 79% of the strains had the blaCTX-M gene. A significant association (p<0.05) was found between the phylogenetic groups, ciprofloxacin resistance, and the age of the patients.
Conclusion: The predominant E. coli phylogroup was B2. We evidenced high resistance to ciprofloxacin and gentamicin, a high proportion of ESBL strains with the blaCTX-M gene, and a significant association between the phylogenetic group and the resistance to ciprofloxacin.
Downloads
References
Organización Panamericana de la Salud. Vigilancia epidemiológica de las infecciones asociadas a la atención de la salud en neonatología. Módulo IV - PAHO; 2013. Fecha de consulta: 18 de febrero de 2022. Disponible en: https://iris.paho.org/handle/10665.2/31361
Yagui-Moscoso M, Vidal-Anzardo M, Rojas-Mezarina L, Sanabria-Rojas H. Prevención de infecciones asociadas a la atención de salud: conocimientos y prácticas en médicos residentes. An Fac Med. 2021;82:131-9. https://doi.org/10.15381/anales.v82i2.19839
Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal Plus. 2018;8:1-32. https://doi.org/10.1128/ecosalplus.ESP-0004-2017
Bonilla-Marciales AP, Chávez-Cañas WO, Hernández-Mogollón RA, Ramón-Jaimes NA. Estrategias de prevención y control de las infecciones en pacientes oncológicos. MedUNAB. 2019;22:356-68.
Velázquez-Brizuela IE, Aranda-Gama J, Camacho-Cortés JL, Ortiz GG. Epidemiología de infecciones nosocomiales en el Instituto Jalisciense de Cancerología. Rev Cuba Salud Pública. 2013;39:19-31.
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58-65. https://doi.org/10.1111/1758-2229.12019
Gonzáles-Rodríguez AO, Pastor HJ, Villafuerte CA, Barrón YL de, Miranda DV de, Cunza SS. Clasificación filogenética de Escherichia coli uropatógena y respuesta inmunometabólica en adultos mayores con infección urinaria en casas de reposo. Arch Med Col. 2019;19:238-46. https://doi.org/10.30554/archmed.19.2.3334.2019
Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543-74. https://doi.org/10.1128/CMR.00125-13
CLSI. M100Ed27 - Performance standards for antimicrobial susceptibility testing. 32nd edition. Fecha de consulta: 21 de febrero de 2022. Disponible en: https://clsi.org/standards/products/microbiology/documents/m100/
Ghasemi Y, Archin T, Kargar M, Mohkam M. A simple multiplex PCR for assessing prevalence of extended-spectrum β-lactamases producing Klebsiella pneumoniae in Intensive Care Units of a referral hospital in Shiraz, Iran. Asian Pac J Trop Med. 2013;6:703-8. https://doi.org/10.1016/S1995-7645(13)60122-4
Chávez-Hidalgo DC. Frecuencia y subtipos del gen blaCTX-M en enterobacterias productoras de BLEE aisladas de urocultivos en el Instituto Nacional de Enfermedades Neoplásicas de enero a diciembre del 2017 [tesis]. Lima: Universidad Nacional Mayor San Marcos; 2019.
Tejada-Llacsa PJ, Huarcaya JM, Melgarejo GC, Gonzáles LF, Cahuana J, Pari RM, et al. Caracterización de infecciones por bacterias productoras de BLEE en un hospital de referencia nacional. An Fac Med. 2015;76:161-6. https://doi.org/10.15381/anales.v76i2.11143
Sader HS, Jones RN, Andrade-Baiocchi S, Biedenbach DJ, SENTRY Participants Group (Latin America). Four-year evaluation of frequency of occurrence and antimicrobial susceptibility patterns of bacteria from bloodstream infections in Latin American medical centers. Diagn Microbiol Infect Dis. 2002;44:273-80. https://doi.org/10.1016/s0732-8893(02)00469-8
Koirala S, Khadka S, Sapkota S, Sharma S, Khanal S, Thapa A, et al. Prevalence of CTX-M β-lactamases producing multidrug resistant Escherichia coli and Klebsiella pneumoniae among patients attending Bir Hospital, Nepal. BioMed Res Int. 2021:9958294. https://doi.org/10.1155/2021/9958294
Dirar MH, Bilal NE, Ibrahim ME, Hamid ME. Prevalence of extended-spectrum β-lactamase (ESBL) and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Enterobacteriaceae isolates from patients in Khartoum, Sudan. Pan Afr Med J. 2020;37:213. https://doi.org/10.11604/pamj.2020.37.213.24988
Demirci M, Ünlü Ö, İstanbullu Tosun A. Detection of O25b-ST131 clone, CTX-M-1 and CTX-M-15 genes via real-time PCR in Escherichia coli strains in patients with UTIs obtained from a university hospital in Istanbul. J Infect Public Health. 2019;12:640-4. https://doi.org/10.1016/j.jiph.2019.02.017
Hernández E, Araque M, Millán Y, Millán B. Prevalencia de b-lactamasa CTX-M-15 en grupos filogenéticos de Escherichia coli uropatógena aisladas en pacientes de la comunidad en Mérida, Venezuela. Invest Clin. 2014;55:13.
Galindo-Méndez M. Caracterización molecular y patrón de susceptibilidad antimicrobiana de Escherichia coli productora de β-lactamasas de espectro extendido en infección del tracto urinario adquirida en la comunidad. Rev Chil Infectol. 2018;35:29-35. https://doi.org/10.4067/s0716-10182018000100029
Gonzáles E, Patiño L, Ore E, Martínez V, Moreno S, Cruzado NB, et al. β-lactamasas de espectro extendido tipo CTX-M en aislamientos clínicos de Escherichia coli y Klebsiella pneumoniae en el Instituto Nacional de Salud del Niño-Breña, Lima, Perú. Rev Médica Hered. 2019;30:242-8. https://doi.org/10.20453/rmh.v30i4.3659
Alsharapy SA, Yanat B, Lopez-Cerero L, Nasher SS, Díaz-De-Alba P, Pascual Á, et al. Prevalence of ST131 clone producing both ESBL CTX-M-15 and AAC(6’)Ib-cr among ciprofloxacin-resistant Escherichia coli isolates from Yemen. Microb Drug Resist Larchmt N. 2018;24:1537-42. https://doi.org/10.1089/mdr.2018.0024
Millán Y, Araque M, Ramírez A. Distribución de grupos filogenéticos, factores de virulencia y susceptibilidad antimicrobiana en cepas de Escherichia coli uropatógena. Rev Chil Infectol. 2020;37:117-23. https://doi.org/10.4067/s0716-10182020000200117
Cole BK, Ilikj M, McCloskey CB, Chávez-Bueno S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS ONE. 2019;14:e0219352. https://doi.org/10.1371/journal.pone.0219352
Ranjbar R, Nazari S, Farahani O. Phylogenetic analysis and antimicrobial resistance profiles of Escherichia coli strains isolated from UTI-suspected patients. Iran J Public Health. 2020;49:1743-9. https://doi.org/10.18502/ijph.v49i9.4094
Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207-17. https://doi.org/10.1038/nrmicro2298
Tsai W-L, Hung C-H, Chen H-A, Wang J-L, Huang I-F, Chiou Y-H, et al. Extended-spectrum β-lactamase-producing Escherichia coli bacteremia: Comparison of pediatric and adult populations. J Microbiol Immunol Infect. 2018;51:723-31. https://doi.org/10.1016/j.jmii.2017.08.005
Some similar items:
- María Fernanda Gutiérrez, Sandra Moreno, Mónica Viviana Alvarado, Andrea Bermúdez, DNA sequence analysis indicates human origin of rotavirus and hepatitis A virus strains from western Colombia , Biomedica: Vol. 29 No. 2 (2009)
- Brian Alejandro Suárez, Claudia Liliana Cuervo, Concepción Judith Puerta, The intergenic region of the histone h2a gene supports two major lineages of Trypanosoma rangeli , Biomedica: Vol. 27 No. 3 (2007)
- Nadia Yadira Castañeda, Jacqueline Chaparro-Olaya, Jaime E. Castellanos, Production and characterization of a polyclonal antibody against rabies virus phosphoprotein , Biomedica: Vol. 27 No. 2 (2007)
- Manuel Muñoz, Juan Carlos Navarro, Mayaro: A re-emerging Arbovirus in Venezuela and Latin America , Biomedica: Vol. 32 No. 2 (2012)
- Andrés Páez, Constanza Nuñez, Clemencia García, Jorge Boshell, Molecular epidemiology of rabies epizootics in Colombia, 1994-2002: evidence of human and canine rabies associated with chiroptera. , Biomedica: Vol. 23 No. 1 (2003)
- Alfonso Benítez-Páez, Sonia Cárdenas-Brito, Mauricio Corredor, Magda Villarroya, María Eugenia Armengod, Impairing methylations at ribosome RNA, a point mutation-dependent strategy for aminoglycoside resistance: The rsmG case , Biomedica: Vol. 34 (2014): Abril, Suplemento 1, Resistencia bacteriana
- Adriana Jiménez, Alejandra Alvarado, Felipe Gómez, Germán Carrero, Claudia Fajardo, Risk factors associated with the isolation of extended spectrum betalactamases producing Escherichia coli or Klebsiella pneumoniae in a tertiary care hospital in Colombia , Biomedica: Vol. 34 (2014): Abril, Suplemento 1, Resistencia bacteriana
- Sandra Liliana Valderrama, Pedro Felipe González, María Alejandra Caro, Natalia Ardila, Beatriz Ariza, Fabián Gil, Carlos Álvarez, Risk factors for hospital-acquired bacteremia due to carbapenem-resistant Pseudomonas aeruginosa in a Colombian hospital , Biomedica: Vol. 36 (2016): Suplemento 1, Microbiología médica
- Elvia Michelli, Adriana Millán, Hectorina Rodulfo, Mirian Michelli, Jesús Luiggi, Numirin Carreño, Marcos de Donato, Identification of enteropathogenic Escherichia coli in children with acute diarrheic syndrome from Sucre State, Venezuela , Biomedica: Vol. 36 (2016): Suplemento 1, Microbiología médica
- Paula Andrea Báez, Carlos Mario Jaramillo, Lina Arismendi, Julio Cesar Rendón, Fabián Cortés-Mancera, Dioselina Peláez, María Mercedes González, Francisco Molina, María Cristina Navas, Evidence of the circulation of hepatitis A virus, subgenotype IA, in environmental samples from Antioquia, Colombia , Biomedica: Vol. 36 (2016): Suplemento 2, Enfermedades virales
Copyright (c) 2022 Biomedica

This work is licensed under a Creative Commons Attribution 4.0 International License.
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























