Análisis de mutaciones en los genes SOD1 y C9orf72 en pacientes con esclerosis lateral amiotrófica, Antioquia, Colombia
Resumen
Introducción. La esclerosis lateral amiotrófica es una enfermedad neurodegenerativa con un posible origen multifactorial, caracterizado por una degeneración progresiva de las neuronas motoras. Hay una gran prevalencia relativa de esta enfermedad en Antioquia; sin embargo, no hay publicaciones de estudios genéticos en Colombia. A pesar de su etiopatogénesis desconocida, hay varios factores de riesgo genético que se encuentran constantemente en el desarrollo de esta enfermedad.
Objetivo. Evaluar las mutaciones G93A y D90A del gen SOD1 y una repetición corta en tándem (Short Tandem Repeat, STR) en el locus C9orf72, en una cohorte de pacientes con esclerosis lateral amiotrófica en Antioquia, Colombia.
Materiales y métodos. Se incluyeron 34 pacientes previamente diagnosticados en el estudio. Una muestra de sangre periférica se usó para extraer el ADN y, posteriormente, genotipificarlo.
Resultados. No se encontraron mutaciones en el gen SOD1 (G93A y D90A), mientras que el C9orf72 exhibe un alelo con una significativa prevalencia en los pacientes del estudio (8 repeticiones del hexanucleótido G4C2).
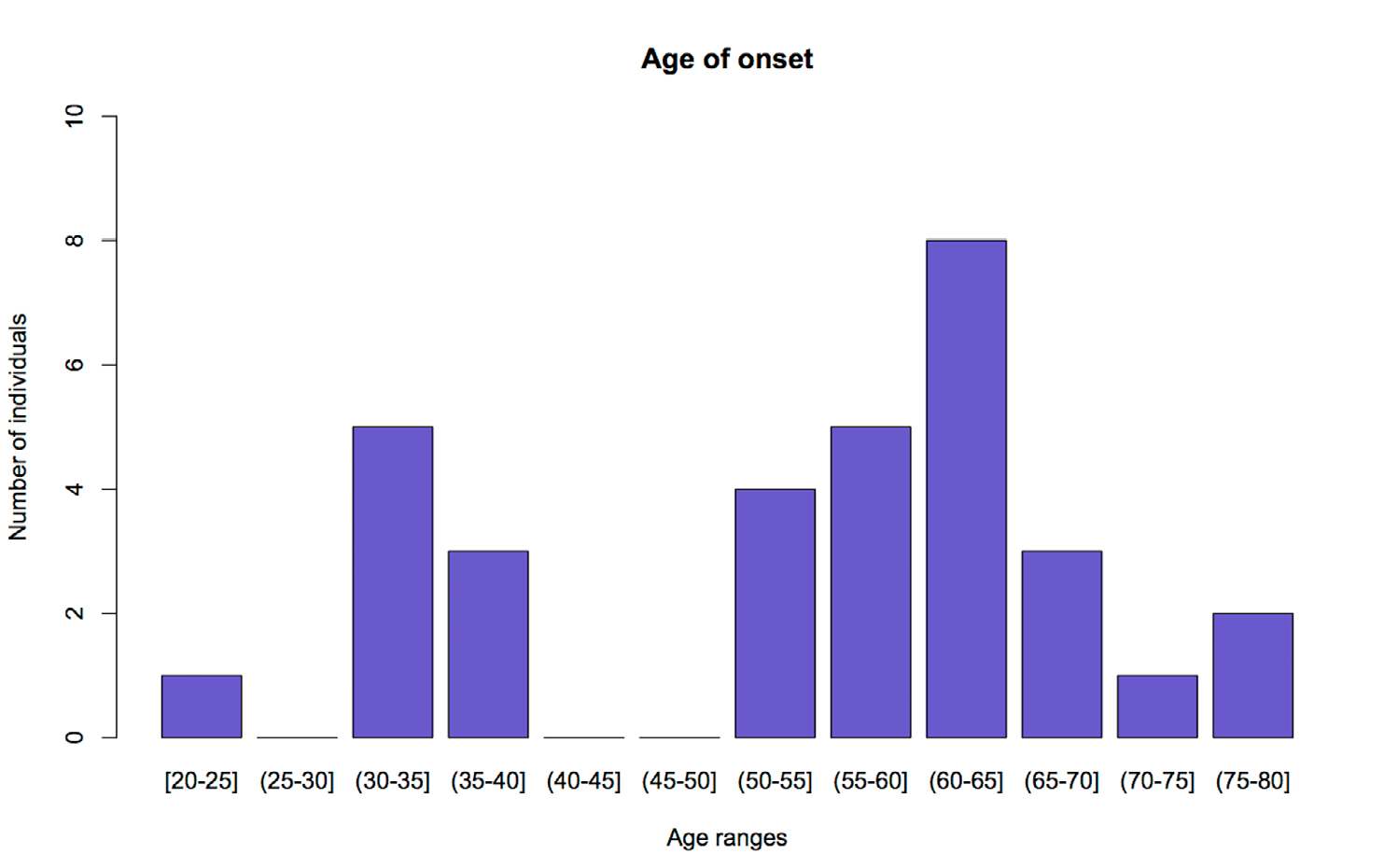
Conclusiones. Se sugiere una asociación entre la repetición en tándem en C9orf72 y la presencia de la esclerosis lateral amiotrófica en la población estudiada. Sin embargo, se sugiere hacer estudios adicionales e incluir un grupo control de la misma población. Además, se detecta un fenómeno de anticipación genética de la enfermedad, dado que los pacientes con el alelo de 8 repeticiones en C9orf72 presentan una edad temprana de aparición de los síntomas.
Descargas
Referencias bibliográficas
Yamashita S, Ando Y. Genotype-phenotype relationship in hereditary amyotrophic lateral sclerosis. Transl Neurodegener. 2015;4:13. https://doi.org/10.1186/s40035-015-0036-y
Turner MR, Verstraete E. What does imaging reveal about the pathology of amyotrophic lateral sclerosis? Curr Neurol Neurosci Rep. 2015;15:45. https://doi.org/10.1007/s11910-015-0569-6
Hamosh A. Online Mendelian Inheritance in Man (OMIM), a knowledge base of human genes and genetic disorders. Nucleic Acids Res. 2004;33:D514-7. https://doi.org/10.1093/nar/gki033
Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124-5. https://doi.org/10.1038/ng0208-124
Rohrer JD, Isaacs AM, Mizielinska S, Mead S, Lashley T, Wray S, et al. C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol. 2015;14:291-301. https://doi.org/10.1016/S1474-4422(14)70233-9
Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J. Rare Dis. 2009;4:3. https://doi.org/10.1186/1750-1172-4-3
Ajroud-Driss S, Siddique T. Sporadic and hereditary amyotrophic lateral sclerosis (ALS). BBA -Mol Basis of Dis. 2015;1852:679-84. https://doi.org/10.1016/j.bbadis.2014.08.010
De Jong SW, Huisman MHB, Sutedja NA, van der Kooi AJ, de Visser M, Schelhaas HJ, et al. Smoking, alcohol consumption, and the risk of amyotrophic lateral sclerosis: A populationbased study. Am J Epidemiol. 2012;176:233-9. https://doi.org/ 10.1093/aje/kws015
Saez-Atienzar S, Bandres-Ciga S, Langston RG, Kim JJ, Choi SW, Reynolds RH, et al. Genetic analysis of amyotrophic lateral sclerosis identifies contributing pathways and cell types. Sci Adv. 2021;7:eabd9036. https://doi.org/10.1126/sciadv.abd9036
Özoğuz A, Uyan Ö, Birdal G, Iskender C, Kartal E, Lahut S, et al. The distinct genetic pattern of ALS in Turkey and novel mutations. Neurobiol Aging. 2015;36:1764.e9-1764.e18. https://doi.org/10.1016/j.neurobiolaging.2014.12.032
Bertolin C, D’Ascenzo C, Querin G, Gaiani A, Boaretto F, Salvoro C, et al. Improving the knowledge of amyotrophic lateral sclerosis genetics: Novel SOD1 and FUS variants. Neurob Aging. 2014;35:1212. http://doi.org/10.1016/j.neurobiolaging.2013.10.093
Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol. 2012;11:232-40. https://doi.org/10.1016/S1474-4422(12)70014-5
Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, et al. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLOS Genet. 2016;12:e1006443. https://doi.org/10.1371/journal.pgen.1006443
Isobe T, Tooi N, Nakatsuji N, Aiba K. Amyotrophic lateral sclerosis models derived from human embryonic stem cells with different superoxide dismutase 1 mutations exhibit differential drug responses. Stem Cell Res. 2015;15:459-68. https://doi.org/10.1016/j.scr.2015.09.006
Mancuso R, Navarro X. Amyotrophic lateral sclerosis: Current perspectives from basic research to the clinic. Prog Neurobiol. 2015;133:1-26. https://doi.org/10.1016/j.pneurobio.2015.07.004
Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-Linked ALSFTD. Neuron. 2011;72:257-68. https://doi.org/10.1016/j.neuron.2011.09.010
Ng ASL, Tan EK. Intermediate C9orf72 alleles in neurological disorders: Does size really matter? J Med Genet. 2017;54:591-7. https://doi.org/10.1136/jmedgenet-2017-104752
Ospina ML, Prieto FE, Pachecho O, Quijano H, Lozano N, Gomez S, et al. Boletín epidemiológico semana 5. 2019. Fecha de consulta: 4 de febrero de 2021. Disponible en: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2019%20Bolet%C3%ADn%20epidemiol%C3%B3gico%20semana%205.pdf
Zapata-Zapata C, Franco-Dáger E, Solano-Atehortúa J, Ahunca-Velásquez L. Esclerosis lateral amiotrófica: actualización. IATREIA. 2016;29:194-205. https://doi.org/10.17533/udea.iatreia.v29n2a08
Zapata-Zapata CH, Dáger EF, Aguirre-Acevedo DC, de Carvalho M, Solano-Atehortua J. Prevalence, incidence, and clinical-epidemiological characterization of amyotrophic lateral sclerosis in Antioquia: Colombia. Neuroepidemiology. 2020;54:251-8. https://doi.org/ 10.1159/000504549
Nikali K, Vanegas JJ, Burley MW, Martínez J, López LM, Bedoya G, et al. Extensive founder effect for distal renal tubular acidosis (dRTA) with sensorineural deafness in an isolated South American population. Am J Med Genet. 2008;146A:2709-12. https://doi.org/ 10.1002/ajmg.a.32495
Carvajal-Carmona LG, Soto ID, Pineda N, Ortiz-Barrientos D, Duque C, Ospina-Duque J, et al. Strong amerind/white sex bias and a possible sephardic contribution among the founders of a population in Northwest Colombia. Am J Hum Genet. 2000;67:1287-95. https://doi.org/10.1016/s0002-9297(07)62956-5
Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61:233-47. https://doi.org/ 10.1034/j.1399-0004.2002.610401.x
De Carvalho M, Dengler R, Eisen A, England JD, Kaji R, Kimura J, et al. Electrodiagnostic criteria for diagnosis of ALS. Clin Neurophysiol. 2008;119:497-503. https://doi.org/ 10.1016/j.clinph.2007.09.143
DeJesús-Hernández M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9orf72 causes chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245-56. https://doi.org/10.1016/j.neuron.2011.09.011
Martín Andrés A. Entry Fisher’s exact and Barnard’s tests. In: Kotz S, Johnson NL, Read CB, editors. Encyclopedia of Statistical Sciences. New York, NY: Wiley-Interscience; 1998. p. 250-8. https://doi.org/10.1002/0471667196.ess0642.pub2
Rousset F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103-6. https://doi.org/10.1111/j.1471-8286.2007.01931.x
Raymond M, Rousset F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248-9. https://doi.org/10.1093/oxfordjournals.jhered.a111573
Couthouis J, Raphael AR, Daneshjou R, Gitler AD. Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet. 2014;10:e1004704. https://doi.org/10.1371/journal.pgen.1004704
Marin B, Boumédiene F, Logroscino G, Couratier P, Babron MC, Leutenegger AL, et al. Variation in worldwide incidence of amyotrophic lateral sclerosis: A meta-analysis. Int J Epidemiol. 2017;46:57-74. https://doi.org/10.1093/ije/dyw061
Chiò A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, et al. Prognostic factors in ALS: A critical review. Amyotroph Lateral Scler. 2009;10:310-23. https://doi.org/10.3109/17482960802566824
Logroscino G, Piccininni M. Amyotrophic lateral sclerosis descriptive epidemiology: The Origin of geographic difference. Neuroepidemiology. 2019;52:93-103. https://doi.org/10.1159/000493386
Carvajal-Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, León P, et al. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet. 2003;112:534-41. https://doi.org/10.1007/s00439-002-0899-8
Chiurazzi P, Oostra BA. Expanding Mutations/Genetic Anticipation. Encyclopedia of Life Sciences. Chichester, UK: John Wiley & Sons, Ltd.; 2006. https://doi.org/10.1038/npg.els.0001463
Suzuki N, Nishiyama A, Warita H, Aoki M. Genetics of amyotrophic lateral sclerosis: Seeking therapeutic targets in the era of gene therapy. J Hum Genet. 2022. https://doi.org/10.1038/s10038-022-01055-8
Byrne S, Heverin M, Elamin M, Walsh C, Hardiman O. Intermediate repeat expansion length in C9orf72 may be pathological in amyotrophic lateral sclerosis. Amyotrop Lateral Scler Frontotemporal Degener. 2014;15:148-50. https://doi.org/10.3109/21678421.2013.838586
Yang Q, Jiao B, Shen L. The development of C9orf72-related amyotrophic lateral sclerosis and frontotemporal dementia disorders. Front Genet. 2020. https://doi.org/10.3389/fgene.2020.562758
Mitteroecker P, Cheverud JM, Pavlicev M. Multivariate analysis of genotype–phenotype association. Genet. 2016;202:1345-63. https://doi.org/10.1534/genetics.115.181339
Namipashaki A, Razaghi-Moghadam Z, Ansari-Pour N. The essentiality of reporting hardyweinberg equilibrium calculations in population-based genetic association studies. Cell J. 2015;17:187-92. https://doi.org/10.22074/cellj.2016.3711
Hedrick PW. Genetics of populations. 4th edition. Sudbury, MA: Jones and Bartlett Publishers; 2011. p. 65-109.
Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N, et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol. 2020;3:526. https://doi.org/10.1038/s42003-020-01251-2
Mesaros M, Lenz S, Lim W, Brown J, Drury L, Roggenbuck J. Investigating the genetic profile of the amyotrophic lateral sclerosis/frontotemporal dementia (ALS-FTD) continuum in patients of diverse race, ethnicity and ancestry. Genes (Basel). 2021;13:76. https://doi.org/10.3390/genes13010076
Štětkářová I, Ehler E. Diagnostics of amyotrophic lateral sclerosis: Up to date. Diagnostics (Basel). 2021;11:231. https://doi.org/10.3390/diagnostics11020231
Huang F, Zhu Y, Hsiao-Nakamoto J, Tang X, Dugas JC, Moscovitch-Lopatin M, et al. Longitudinal biomarkers in amyotrophic lateral sclerosis. Ann Clin Transl Neurol. 2020;7:1103-16. https://doi.org/10.1002/acn3.51078
Álvarez-Fernández A, Bernal MJ, Fradejas I, Martín Ramírez A, Md Yusuf NA, Lanza M, et al. KASP: A genotyping method to rapid identification of resistance in Plasmodium falciparum. Malar J. 2021;20:16. https://doi.org/10.1186/s12936-020-03544-7
Algunos artículos similares:
- Brian Alejandro Suárez, Claudia Liliana Cuervo, Concepción Judith Puerta, La región intergénica del gen H2A apoya las subpoblaciones KP1(-) y KP1(+) de Trypanosoma rangeli , Biomédica: Vol. 27 Núm. 3 (2007)
- Patricia Escandón, Popchai Ngamskulrungroj, Wieland Meyer, Elizabeth Castañeda, Determinación in vitro de la pareja sexual en aislamientos del complejo Cryptococcus neoformans , Biomédica: Vol. 27 Núm. 2 (2007)
- Andrea Gómez, Gustavo Salguero, Herbert García, Fabio Aristizábal, Oscar Gutiérrez, Luis Alberto Angel, Jorge Padrón, Carlos Martínez, Humberto Martínez, Omar Malaver, Rosa Barvo, Alejandro Giraldo, Detección de mutaciones de los genes hMLH1 y hMSH2 del sistema de reparación de malos apareamientos del ADN en familias colombianas sospechosas de cancer colorrectal no polipósico hereditario (síndrome de Lynch). , Biomédica: Vol. 25 Núm. 3 (2005)
- Diana León-Luna , Alexander Fajardo-Loyola , José Yareta-Yareta , Antonio Burgos-Espejo , Carlos Peralta-Siesquen , Marco Galarza-Pérez , Pool Marcos-Carbajal, Caracterización molecular de enterobacterias multirresistentes en dos departamentos de la selva peruana , Biomédica: Vol. 41 Núm. Sp. 2 (2021): Octubre, Infecciones bacterianas y virales
Derechos de autor 2022 Biomédica

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
| Estadísticas de artículo | |
|---|---|
| Vistas de resúmenes | |
| Vistas de PDF | |
| Descargas de PDF | |
| Vistas de HTML | |
| Otras vistas | |