Analysis of hepatitis B virus genotypes by restriction fragment length polymorphism
Abstract
Introduction: Ten viral genotypes (A-J) distributed in all continents have been described for hepatitis B virus (HBV). One of the methodologies for determining the viral genotype is the restriction fragment length polymorphism (RFLP) technique, a simple and relatively inexpensive method, albeit with some limitations.
Objective: The initial objective of the project was to identify the HBV genotypes by RFLP in serum samples obtained from patients and blood donors. However, due to the discrepancies of RFLP patterns it was also necessary to perform phylogenetic genotyping and in silico analysis of HBV sequences.
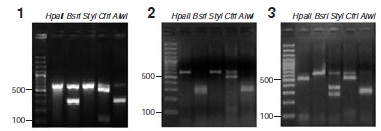
Materials and methods: We obtained 56 serum samples. DNA extraction was followed by PCR amplification of a fragment of HBV ORF S. We analyzed PCR products by RFLP with AlwI, BsrI, CfrI, HpaII and StyI, and we sequenced some. We compared the patterns obtained with those in previous reports. We also performed RFLP analysis in silico since we found differences between the patterns expected and those obtained
Results: We identified genotypes A and F, subgenotype F3, in the samples. This result is in agreement with those of previous studies carried out in Colombia; indeed, subgenotype F3 is the most frequent in the Andean region of the country, while genotype A is the most frequent HBV genotype in the western region (department of Chocó). Based on the in silico analysis of 229 HBV sequences from GenBank and 11 sequences of this study, we identified the RLFP pattern for genotype F, subgenotype F3, and we described some modifications of genotype A RFLP patterns.
Conclusions: We identified the single nucleotide polymorphism pattern for genotype F, subgenotype F3, by in silico analysis and sequencing. Further robust in silico analyses are necessary to validate the RFLP patterns of HBV genotype and subgenotypes.
Downloads
References
World Health Organization. Hepatitis B. Date of entry: April 15, 2015. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/
Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337: 1733-45. http://dx.doi.org/10.1056/NEJM199712113372406
Milich D, Liang TJ. Exploring the biological basis of hepatitis B antigen in hepatitis B virus infection. Hepatol Baltim Md. 2003;38:1075-86. http://dx.doi.org/10.1053/jhep. 2003.50453
Heise T, Sommer G, Reumann K, Meyer I, Will H, Schaal H. The hepatitis B virus PRE contains a splicing regulatory element. Nucleic Acids Res. 2006;34:353-63. http://dx.doi.org/10.1093/nar/gkj440
Norder H, Couroucé A-M, Coursaget P, Echevarría JM, Lee S-D, Mushahwar IK, et al. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. http://dx.doi.org/10.1159/000080872
Campos RH, Mbayed VA, Piñeiro Y Leone FG. Molecular epidemiology of hepatitis B virus in Latin America. J Clin Virol. 2005;34(Suppl.2):S8-13. http://dx.doi.org/10.1016/S1386-6532(05)80028-9
Panduro A, Maldonado-González M, Fierro NA, Román S. Distribution of HBV genotypes F and H in México and Central America. Antivir Ther. 2013;18:475-84. http://dx.doi.org/10.3851/IMP2605
Alvarado-Mora MV, Pinho JR. Distribution of HBV geno-types in Latin America. Antivir Ther. 2013;18:459-65. http://dx.doi.org/10.3851/IMP2599
Sunbul M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J Gastroenterol. 2014;20: 5427-34. http://dx.doi.org/10.3748/wjg.v20.i18.5427
Lin CL, Kao JH. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436. http://dx.doi.org/10.1101/cshperspect.a021436
Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489-503. http://dx. doi.org/10.1006/viro.1994.1060
Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutiérrez MF, Carrilho FJ, Pinho JR. Molecular epidemiology and genetic diversity of hepatitis B virus genotype E in an isolated Afro-Colombian community. J Gen Virol. 2010;91:501-8. http://dx.doi.org/10.1099/vir.0. 015958-0
Olinger CM, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, et al. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14:1777–80. http://dx.doi.org/10.3201/eid1411. 080437
Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538-47. http://dx.doi.org/10.1128/JVI.00462-09
Gandhi MJ, Yang GG, McMahon BJ, Vyas GN. Hepatitis B virions isolated with antibodies to the pre-S1 domain reveal occult viremia by PCR in Alaska Native HBV carriers who have seroconverted. Transfusion. 2000;40:910-6. http://dx. doi.org/10.1046/j.1537-2995.2000.40080910.x
Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: A new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-73. http://dx.doi.org/10.1099/0022-1317-83-8-2059
Devesa M, Pujol FH. Hepatitis B virus genetic diversity in Latin America. Virus Res. 2007;127:177-84. http://dx.doi.org/10.1016/j.virusres.2007.01.004
Zehender G, Ebranati E, Gabanelli E, Sorrentino C, Lo Presti A, Tanzi E, et al. Enigmatic origin of hepatitis B virus: An ancient travelling companion or a recent encounter? World J Gastroenterol. 2014;20:7622-34. http://dx.doi.org/10. 3748/wjg.v20.i24.7622
Alvarado-Mora MV, Gutiérrez-Fernández MF, Gomes-Gouvêa MS, de Azevedo-Neto RS, Carrilho FJ, Pinho JR. Hepatitis B (HBV), hepatitis C (HCV) and hepatitis delta (HDV) viruses in the Colombian population –How Is the epidemiological situation? PLoS One. 2011;6:e18888. http://dx.doi.org/10.1371/journal.pone.0018888
Instituto Nacional de Salud. Boletín Epidemiológico semanal: semana 53 de 2014. Date of entry: June 2, 2015. Available from: http://www.ins.gov.co/boletin-epidemiologico/Boletn%20Epidemiolgico/2014%20Boletin%20epidemiologico%20semana%2053.pdf
Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B Virus by homology in nucleotide sequence: Comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-83. http://dx. doi.org/10.1099/0022-1317-69-10-2575
Vernet G, Tran N. The DNA-Chip technology as a new molecular tool for the detection of HBV mutants. J Clin Virol. 2005;34 (Suppl.1):S49-53. http://dx.doi.org/10.1016/S1386-6532(05)80010-1
Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, et al. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66-71. http://dx.doi.org/10.1016/S0014-5793(99)00471-8
Naito H, Hayashi S, Abe K. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J Clin Microbiol. 2001;39:362-4. http://dx.doi.org/10.1128/JCM.39. 1.362-364.2001
Usuda S, Okamoto H, Iwanari H, Baba K, Tsuda F, Miyakawa Y, et al. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods. 1999;80:97-112. http://dx.doi.org/10.1016/S0166-0934(99)00039-7
Osiowy C, Giles E. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J Clin Microbiol. 2003;41:5473-7. http://dx.doi.org/10.1128/JCM.41.12.5473-5477.2003
Lindh M, Andersson AS, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus--Large-scale analysis using a new genotyping method. J Infect Dis. 1997;175:1285-93. http://dx.doi.org/10.1086/516458
Lindh M, González JE, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163-74. http://dx.doi.org/10.1016/S0166-0934(98)00026-3
Zeng GB, Wen SJ, Wang ZH, Yan L, Sun J, Hou JL. A novel hepatitis B virus genotyping system by using restriction fragment length polymorphism patterns of S gene amplicons. World J Gastroenterol. 2004;10:3132-6. http://dx.doi.org/10.3748/wjg.v10.i21.3132
Venegas M, Muñoz G, Hurtado C, Álvarez L, Velasco M, Villanueva RA, et al. Prevalence of hepatitis B virus genotypes in chronic carriers in Santiago, Chile. Arch Virol. 2008;153:2129-32. http://dx.doi.org/10.1007/s00705-008-0231-6
Gulube Z, Chirara M, Kew M, Tanaka Y, Mizokami M, Kramvis A. Molecular characterization of hepatitis B virus isolates from Zimbabwean blood donors. J Med Virol. 2011;83:235-44. http://dx.doi.org/10.1002/jmv.21954
Ouneissa R, Bahri O, Ben Yahia A, Touzi H, Azouz MM, Ben Mami N, et al. Evaluation of PCR-RFLP in the pre-S region as molecular method for hepatitis B virus genotyping. Hepat Mon. 2013;13:e11781. http://dx.doi.org/10.5812/hepatmon.11781
Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95-8.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731-9. http://dx.doi.org/10.1093/molbev/msr121
Nabuco LC, Mello FC, Gomes S de A, Perez RM, Soares JA, Coelho HS, et al. Hepatitis B virus genotypes in Southeast Brazil and its relationship with histological features. Mem Inst Oswaldo Cruz. 2012;107:785-9. http://dx.doi.org/10.1590/S0074-02762012000600013
Gopalakrishnan D, Keyter M, Shenoy KT, Leena KB, Thayumanavan L, Thomas V, et al. Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J Gastroenterol. 2013;19:9294-306. http://dx.doi.org/10.3748/wjg.v19.i48.9294
Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82:5657-63. http://dx.doi.org/10.1128/JVI.02556-07
Pourkarim MR, Amini-Bavil-Olyaee S, Lemey P, Maes P, van Ranst M. Are hepatitis B virus “subgenotypes” defined accurately? J Clin Virol. 2010;47:356-60. http://dx.doi.org/10.1016/j.jcv.2010.01.015
Hübschen JM, Mbah PO, Forbi JC, Otegbayo JA, Olinger CM, Charpentier E, et al. Detection of a new subgenotype of hepatitis B virus genotype A in Cameroon but not in neighbouring Nigeria. Clin Microbiol Infect. 2011;17:88-94. http://dx.doi.org/10.1111/j.1469-0691.2010.03205.x
Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9. Arch Virol. 2011;156:855-68. http://dx.doi.org/10.1007/s00705-011-0926-y
Mulyanto, Pancawardani P, Depamede SN, Wahyono A, Jirintai S, Nagashima S, et al. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012;163:129-40. http://dx.doi.org/10.1016/j.virusres.2011.09.002
Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Dény P, et al. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609-20. http://dx.doi.org/10.1099/vir.0.018127-0
Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutiérrez MF, Carrilho FJ, Pinho JR. Dynamics of hepatitis D (delta) virus genotype 3 in the Amazon region of South America. Infect Genet Evol. 2011;11:1462-8. http://dx.doi.org/10.1016/j.meegid.2011.05.020
Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutiérrez MF, Carrilho FJ, Pinho JR. Phylogenetic analy-sis of complete genome sequences of hepatitis B virus from an Afro-Colombian community: Presence of HBV F3/A1 recombinant strain. Virol J. 2012;9:244. http://dx.doi.org/10.1186/1743-422X-9-244
Bautista-Amorocho H, Castellanos-Domínguez YZ, Rodríguez-Villamizar LA, Velandia-Cruz SA, Becerra-Peña JA, Farfán-García AE. Epidemiology, risk factors and genotypes of HBV in HIV-infected patients in the Northeast region of Colombia: High prevalence of occult hepatitis B and F3 subgenotype dominance. PLoS One. 2014;9: e114272. http://dx.doi.org/10.1371/journal.pone.0114272
Cortés-Mancera F, Loureiro CL, Hoyos S, Restrepo JC, Correa G, Jaramillo S, et al. Etiology and viral genotype in patients with end-stage liver diseases admitted to a hepatology unit in Colombia. Hepat Res Treat. 2011;2011:363205. http://dx.doi.org/10.1155/2011/363205
Devesa M, Loureiro CL, Rivas Y, Monsalve F, Cardona N, Duarte MC, et al. Subgenotype diversity of hepatitis B virus American genotype F in Amerindians from Venezuela and the general population of Colombia. J Med Virol. 2008;80:20-6. http://dx.doi.org/10.1002/jmv.21024
Ríos-Ocampo WA, Cortés-Mancera F, Olarte JC, Soto A, Navas MC. Occult hepatitis B virus infection among blood donors in Colombia. Virol J. 2014;11:206. http://dx.doi.org/10.1186/s12985-014-0206-z
Ríos D, di Filippo D, Insuasty M, Rendón JC, Ríos WA, Medina C, et al. Hepatitis B infections in individuals with exposure factors in Quibdó and Apartadó, Colombia. Rev Col Gastroenterol. 2015;30:11-8.
Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutiérrez MF, Botelho L, Carrilho FJ, et al. Molecular characterization of the hepatitis B virus genotypes in Colombia: A Bayesian inference on the genotype F. Infect Genet Evol. 2011;11:103-8. http://dx.doi.org/10.1016/j.meegid.2010.10.003
di Filippo Villa D, Cortés-Mancera F, Payares E, Montes N, de la Hoz F, Arbeláez MP, et al. Hepatitis D virus and hepatitis B virus infection in Amerindian communities of the Amazonas state, Colombia. Virol J. 2015;12:172. http://dx.doi.org/10.1186/s12985-015-0402-5
Departamento Administrativo Nacional de Estadística (DANE). Censo general. Bogotá, D.C.: DANE; 2005.
Quintero A, Martínez D, Alarcón De Noya B, Costagliola A, Urbina L, González N, et al. Molecular epidemiology of hepatitis B virus in Afro-Venezuelan populations. Arch Virol. 2002;147:1829-36. http://dx.doi.org/10.1007/s00705-002-0842-2
Motta-Castro AR, Martins RM, Yoshida CF, Teles SA, Paniago AM, Lima KM, et al. Hepatitis B virus infection in isolated Afro-Brazilian communities. J Med Virol. 2005;77: 188-93. http://dx.doi.org/10.1002/jmv.20435
Martinot-Peignoux M, Lapalus M, Laouénan C, Lada O, Netto-Cardoso AC, Boyer N, et al. Prediction of disease reactivation in asymptomatic hepatitis B e antigen-negative chronic hepatitis B patients using baseline serum measurements of HBsAg and HBV-DNA. J Clin Virol. 2013;58:401-7. http://dx.doi.org/10.1016/j.jcv.2013.08.010
Gupta E, Kumar A, Choudhary A, Kumar M, Sarin SK. Serum hepatitis B surface antigen levels correlate with high serum HBV DNA levels in patients with chronic hepatitis B: A cross-sectional study. Indian J Med Microbiol. 2012;30:150-4. http://dx.doi.org/10.4103/0255-0857.96664
Portilho MM, Baptista ML, da Silva M, de Sousa PS, Lewis-Ximenez LL, Lampe E, et al. Usefulness of in-house PCR methods for hepatitis B virus DNA detection. J Virol Methods. 2015;223:40-4. http://dx.doi.org/10.1016/j.jviromet.2015.07.010
Some similar items:
- Efren Avendaño-Tamayo, Omer Campo, Juan Camilo Chacón-Duque, Ruth Ramírez, Winston Rojas, Piedad Agudelo-Flórez, Gabriel Bedoya, Berta Nelly Restrepo, Variants in the TNFA, IL6 and IFNG genes are associated with the dengue severity in a sample from Colombian population , Biomedica: Vol. 37 No. 4 (2017)
- Clemencia Ovalle-Bracho, Carolina Camargo, Yira Díaz-Toro, Marcela Parra-Muñoz, Molecular typing of Leishmania (Leishmania) amazonensis and species of the subgenus Viannia associated with cutaneous and mucosal leishmaniasis in Colombia: A concordance study , Biomedica: Vol. 38 No. 1 (2018)
- Liliana Franco-Hincapié, Constanza Elena Duque, María Victoria Parra, Natalia Gallego, Alberto Villegas, Andrés Ruiz-Linares, Gabriel Bedoya, Association between polymorphism in uncoupling proteins and type 2 diabetes in a northwestern Colombian population , Biomedica: Vol. 29 No. 1 (2009)
- Ana Margarita Montalvo, Lianet Monzote, Jorge Fraga, Ivón Montano, Carlos Muskus, Marcel Marín, Simonne De Donck, Iván Darío Vélez, Jean Claude Dujardin, PCR-RFLP and RAPD for typing neotropical Leishmania , Biomedica: Vol. 28 No. 4 (2008)
- Dioselina Peláez-Carvajal, Nidia Janeth Forero, Martha Escalante-Mora, Katherine Laiton-Donato, José Aldemar Usme-Ciro, Genetic variability in coding regions of the surface antigen and reverse transcriptase domain of hepatitis B virus polymerase, Colombia, 2002-2014 , Biomedica: Vol. 38 No. Sup. 2 (2018): Suplemento 2, Medicina tropical
- María Teresa Arango, Carlos Jaramillo, María Camila Montealegre, Mabel Helena Bohórquez, María del Pilar Delgado, Genetic characterization of the interleukin 1 β polymorphisms -511, -31 y +3954 in a Colombian population with dyspepsia , Biomedica: Vol. 30 No. 2 (2010)
- Ana M. Montalvo, Jorge Fraga, Ivón Montano, Lianet Monzote, Gert Van der Auwera, Marcel Marín, Carlos Muskus, Molecular identification of Leishmania spp. clinical isolates from Colombia based on hsp70 gene , Biomedica: Vol. 36 (2016): Suplemento 1, Microbiología médica
- Diana María Valencia, Carlos Andrés Naranjo, María Victoria Parra, María Antonieta Caro, Ana Victoria Valencia, Carlos José Jaramillo, Gabriel Bedoya, Association and interaction of AGT, AGTR1, ACE, ADRB2, DRD1, ADD1, ADD2, ATP2B1, TBXA2R and PTGS2 genes on the risk of hypertension in Antioquian population , Biomedica: Vol. 33 No. 4 (2013)
- Diana Carolina López, Carlos Jaramillo, Felipe Guhl, Population structure and genetic variability of Rhodnius prolixus (Hemiptera: Reduviidae) from different geographic areas of Colombia. , Biomedica: Vol. 27 No. 1esp (2007): Enfermedad de Chagas
- Alberto Gómez, Sandra J. Ávila, Ignacio Briceño, Correlation analysis of surnames and Y-chromosome genetic heritage in 3 provinces of southwestern Colombia , Biomedica: Vol. 28 No. 3 (2008)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























