High-resolution melting analysis based on specific genomic regions: A promising tool for the diagnosis and typing of species causing cutaneous leishmaniasis in Colombia
Abstract
Introduction: Cutaneous leishmaniasis, caused by parasites of the genus Leishmania, is a disease with high incidence in Colombia. The diagnosis and identification of the infectious species are critical factors when selecting and initiating treatment. Currently, the methods for diagnosing and typing cutaneous leishmaniasis require complicated procedures and there is a need for the validation of new molecular markers and methods to simplify the process.
Objective: To develop a tool based in PCR melting curves (PCR-HRM) for the diagnosis and typing of the three Leishmania species of epidemiological importance for cutaneous leishmaniasis in Colombia.
Materials and methods: The genomes of Leishmania panamensis, L. braziliensis and L. guyanensis were compared with bioinformatic methods. The species-specific regions were then validated using PCR. For the selected markers, a PCR-HRM was designed, and validity and security parameters were estimated using isolates from Colombian patients previously characterized by PCR-RFLP of the hsp70 gene.
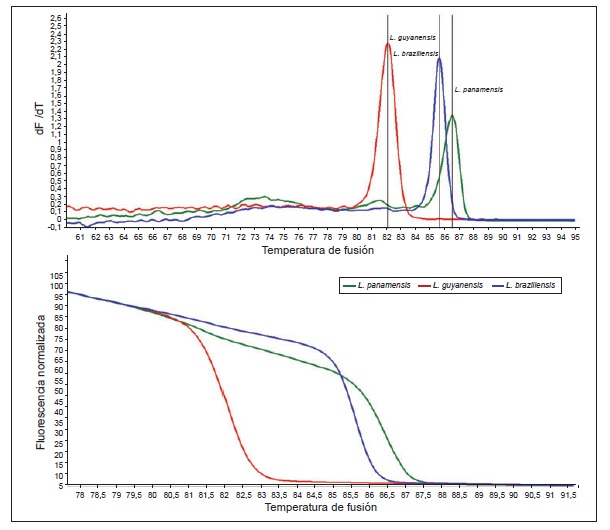
Results: The comparative genomic analysis yielded 24 species-specific regions. However, the PCR validation identified only one marker that was specific to each Leishmania species. The other markers showed cross amplification. The detection limit for the three selected markers was one parasite. The sensitivity, specificity, predictive positive and negative values were 91.4%, 100%, 100% and 75%, respectively.
Conclusions: The three selected regions can be used as molecular markers in the diagnosis and typing of the causative species of cutaneous leishmaniasis in Colombia.
Downloads
References
Mcgwire BS, Satoskar AR. Leishmaniasis: Clinical syndro-mes and treatment. QJM. 2014;107: 7-14. https://doi.or/10. 1093/qjmed/hct116
Reithinger R, Dujardin JC. Molecular diagnosis of leish-maniasis: Current status and future applications. J Clin Micro-biol. 2007;45: 21-5. https://doi.or/10.1128/Jcm.02029-06
World Health Organization. Control of the leishmaniases. Report of a meeting of the WHO Expert Committee on the Control of Leishmaniases. Geneva: WHO; 2010.
Pigott DM, Bhatt S, Golding N, Duda KA, Battle KE, Brady OJ, et al. Global distribution maps of the Leishmaniases. Elife. 2014;3:e02851 https://doi.or/10.7554/eLife.02851.001
Dantas Brito M, Campilho F, Branca R, Pinho Vaz C, Silva C, Sousa T, et al. Visceral leishmaniasis: A differential diagnosis to remember after bone marrow transplantation. Case Rep Hematol. 2014;2014:587912. https://doi.or/10. 1155/2014/587912
Rodríguez-Brito S, Camacho E, Mendoza M, Niño-Vega GA. Differential identification of Sporothrix spp. and Leishmania spp. by conventional PCR and qPCR in multiplex format. Med Mycol. 2014;53: 22-7. https://doi.or/10.1093/mmy/myu065
Fernández OL, Díaz-Toro Y, Ovalle C, Valderrama L, Muvdi S, Rodríguez I, et al. Miltefosine and antimonial drug susceptibility of Leishmania viannia species and populations in regions of high transmission in Colombia. PLoS Negl Trop Dis. 2014;8:e2871. https://doi.or/10.1371/journal.pntd.0002871
Aït-Oudhia K, Gazanion E, Vergnes B, Oury B, Sereno D. Leishmania antimony resistance: What we know what we can learn from the field. Parasitol Res. 2011;109: 1225-32. https://doi.or/10.1007/s00436-011-2555-5
Tuon F, Amato V, Graf M, Siqueira A. Treatment of New World cutaneous leishmaniasis -a systematic review with a meta-analysis. Int J Dermatol. 2008;47:109-24. https://doi.or/10.1111/j.1365-4632.2008.03417.x
Van der Auwera G, Dujardina JC. Species typing in dermal leishmaniasis. Clin Microbiol Rev. 2015;28:265-94.https://doi.or/10.1128/CMR.00104-14
Goto H, Lindoso JA. Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev Anti Infect Ther. 2010;8:419-33. https://doi.or/10.1586/eri. 10.19
Ben Abda I, De Monbrison F, Bousslimi N, Aoun K, Bouratbine A, Picot S. Advantages and limits of real-time PCR assay and PCR-restriction fragment length poly-morphism for the identification of cutaneous Leishmania species in Tunisia. Trans R Soc Trop Med Hyg. 2011;105:17–22. https://doi.or/10.1016/j.trstmh.2010.09.003
Montalvo AM, Fraga J, Maes I, Dujardin J-C, Van Der Auwera G. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012;31:1453-61. https://doi.or/10.1007/s10096-011-1463-z
González-Marcano E, Kato H, Concepción JL, Márquez ME, Mondolfi AP. Polymerase chain reaction diagnosis of leishmaniasis: A species-specific approach. Methods Mol Biol. 2016;1392:113-24. https://doi.or/10.1007/978-1-4939-3360-0_11
Monroy-Ostria A, Nasereddin A, Monteon VM, Guzmán-Bracho C, Jaffe CL. ITS1 PCR-RFLP diagnosis and charac-terization of leishmania in clinical samples and strains from cases of human cutaneous leishmaniasis in states of the Mexican Southeast. Interdiscip Perspect Infect Dis. 2014;2014:607287. https://doi.or/10.1155/2014/607287
Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436-42. https://doi.or/10.1126/science.1112680
Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. https://doi.or/10.1186/1471-2164-6-127
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. https://doi.or/10.1186/gb-2004-5-2-r12
Clark JM, Joyce CM, Beardsley GP. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol. 1987;198:123-7. https://doi.or/10.1016/0022-2836(87)90462-1
Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, et al. Heat-shock protein 70 PCR-RFLP: A universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010;137:1159-68. https://doi.or/10.1017/S0031182010000089
Montalvo AM, Fraga J, El Safi S, Gramiccia M, Jaffe CL, Dujardin JC, et al. Direct Leishmania species typing in Old World clinical samples: Evaluation of 3 sensitive methods based on the heat-shock protein 70 gene. Diagn Microbiol Infect Dis. 2014;80:35-9. https://doi.or/10.1016/j.diagmicrobio.2014.05.012
Cruz-Barrera ML, Ovalle-Bracho C, Ortegon-Vergara V, Pérez-Franco JE, Echeverry MC. Improving Leishmania species identification in different types of samples from cutaneous lesions. J Clin Microbiol. 2015;53:1339-41. https://doi.or/10.1128/JCM.02955-14
Bangdiwala SI, Haedo AS, Natal ML, Villaveces A. The agreement chart as an alternative to the receiver-operating characteristic curve for diagnostic tests. J Clin Epidemiol. 2008;61:866-74. https://doi.or/10.1016/j.jclinepi. 2008.04.002
Simel DL, Samsa GP, Matchar DB. Likelihood ratios with confidence: Sample size estimation for diagnostic test studies. J Clin Epidemiol. 1991;44:763-70. https://doi.or/10. 1016/0895-4356(91)90128-V
Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561-77. https://doi.or/10. 1016/S0140-6736(05)67629-5
Ponce C, Ponce E, Cruz A, Kreutzer R, McMahon-Pratt D, Neva F. Leishmania donovani chagasi: New clinical variant of cutaneous leishmaniasis in Honduras. Lancet. 1991;337:67-70.
Kebede N, Oghumu S, Worku A, Hailu A, Varikuti S, Satoskar AR. Multilocus microsatellite signature and identification of specific molecular markers for Leishmania aethiopica. Parasit Vectors. 2013;6:160. https://doi.or/10. 1186/1756-3305-6-160
Leelayoova S, Siripattanapipong S, Hitakarun A, Kato H, Tan-ariya P, Siriyasatien P, et al. Multilocus characteri-zation and phylogenetic analysis of Leishmania siamensis isolated from autochthonous visceral leishmaniasis cases, southern Thailand. BMC Microbiol. 2013;13:60. https://doi.or/10.1186/1471-2180-13-60
Bhattacharyya R, Das K, Sen S, Roy S, Majumder HK. Development of a genus specific primer set for detection of Leishmania parasites by polymerase chain reaction. FEMS Microbiol Lett. 1996;135:195-200.
Mathis A, Deplazes P. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J Clin Microbiol. 1995;33:1145-9.
Cavalcanti MD, Dantas-Torres F, de Albuquerque SDG, de Morais RC, de Brito ME, Otranto D, et al. Quantitative real time PCR assays for the detection of Leishmania (Viannia) braziliensis in animals and humans. Mol Cell Probes. 2013;27:122-8. https://doi.or/10.1016/j.mcp.2013.01.003
Liew M, Nelson L, Margraf R, Mitchell S, Erali M, Mao R, et al. Genotyping of human platelet antigens 1 to 6 and 15 by high-resolution amplicon melting and conventional hybridization probes. J Mol Diagn. 2006;8:97-104. https://doi.or/10.2353/jmoldx.2006.050053
Tsukayama P, Núñez JH, De Los Santos M, Soberón V, Lucas CM, Matlashewski G, et al. A FRET-Based Real-Time PCR assay to identify the main causal agents of New World tegumentary leishmaniasis. PLoS Negl Trop Dis. 2013;7:21956. https://doi.or/10.1371/journal.pntd.0001956
Chiaramonte MG, Zwirner NW, Caropresi SL, Taranto NJ, Malchiodi EL. Trypanosoma cruzi and Leishmania spp. human mixed infection. Am J Trop Med Hyg. 1996;54:271-3. https://doi.org/10.4269/ajtmh.1996.54.271
Saldarriaga OA, Castellanos-González A, Porrozzi R, Baldeviano GC, Lescano AG, de Los Santos MB, et al. An innovative field-applicable molecular test to diagnose cutaneous Leishmania viannia spp. infections. PLoS Negl Trop Dis. 2016;10:e0004638. https://doi.or/10.1371/journal.pntd.0004638
Mary C, Faraut F, Lascombe L, Dumon H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol. 2004;42:5249-55. https://doi.or/10.1128/JCM.42.11.5249-5255.2004
Some similar items:
- Jazzmín Arrivillaga-Henríquez, Sandra Enríquez, Vanessa Romero, Gustavo Echeverría, Jorge Pérez-Barrera, Ana Poveda, Juan-Carlos Navarro, Alon Warburg, Washington Benítez, Eco-epidemiological aspects, natural detection and molecular identification of Leishmania spp. in Lutzomyia reburra, Lutzomyia barrettoi majuscula and Lutzomyia trapidoi , Biomedica: Vol. 37 No. Sup. 2 (2017): Suplemento 2, Entomología médica, 2017
- Camilo Andrés Morales, Juliana Palacio, Gerzaín Rodríguez, Yenny Carolina Camargo, Zosteriform cutaneous leishmaniasis due to Leishmania (Viannia) panamensis and Leishmania (Viannia) braziliensis: Report of three cases , Biomedica: Vol. 34 No. 3 (2014)
- Luis Alberto Cortés, Jhon James Fernández, Species of Lutzomyia involved in an urban focus of visceral and cutaneous leishmaniasis , Biomedica: Vol. 28 No. 3 (2008)
- Clemencia Ovalle-Bracho, Carolina Camargo, Yira Díaz-Toro, Marcela Parra-Muñoz, Molecular typing of Leishmania (Leishmania) amazonensis and species of the subgenus Viannia associated with cutaneous and mucosal leishmaniasis in Colombia: A concordance study , Biomedica: Vol. 38 No. 1 (2018)
- Olga Lucía Cabrera, Laureano Mosquera, Erika Santamaría, Sand flies (Diptera: Psychodidae) of Guaviare Province, Colombia, with 4 new records for the country , Biomedica: Vol. 29 No. 1 (2009)
- Diego Fernando Zea, Martín Prager, Roger Adrian Figueroa, María Consuelo Miranda, Mucosal complication of cutaneous leishmaniasis , Biomedica: Vol. 29 No. 1 (2009)
- Marcel Marín, Yudy Alexandra Aguilar, José Robinson Ramírez, Omar Triana, Carlos Enrique Muskus, Molecular and immunological analyses suggest the absence of hydrophilic surface proteins in Leishmania (Viannia) panamensis , Biomedica: Vol. 28 No. 3 (2008)
- Eduar Elías Bejarano, Diana Sierra, Iván Darío Vélez, New findings on the geographic distribution of the verrucarum group (Diptera: Psychodidae) in Colombia. , Biomedica: Vol. 23 No. 3 (2003)
- María Angélica Contreras, Rafael José Vivero, Eduar Elías Bejarano, Lina María Carrillo, Iván Darío Vélez, New records of phlebotomine sand flies (Diptera: Psychodidae) near the Amoya River in Chaparral, Tolima , Biomedica: Vol. 32 No. 2 (2012)
- Sandra Milena Barrera, Manuel Alberto Pérez, Angélica Knudson, Rubén Santiago Nicholls, Ángela Patricia Guerra, Genotypic survery of Plasmodium falciparum based on the msp1, msp2 and glurp genes by multiplex PCR , Biomedica: Vol. 30 No. 4 (2010)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























