Importance of determining variations in the number of copies in newborns with autosomal aneuploidies
Abstract
Introduction: Aneuploidies are frequent genetic disorders in clinical practice. However, little is known about other genetic variants that may influence the final phenotype.
Objective: To determine the variations in the number of copies and regions with homozygosity greater than 0.5% or larger than 10 Mb in newborns with autosomal aneuploidies.
Materials and methods: We performed a chromosomal microarray analysis on newborns with autosomal aneuploidies (n=7), trisomy 21 (n=5), and trisomy 18 (n=2) evaluated at the Hospital Antonio Lorena and Hospital Regional of Cusco, Perú, during 2018.
Results: We found pathogenic and probably pathogenic variants in the number of copies in other genomic regions different to chromosomes 21 or 18 in two neonates. Additionally, we found two variants bigger than 500 kpb of unknown pathogenicity.
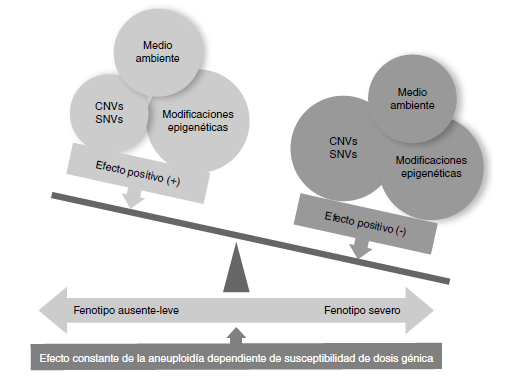
Conclusions: Although the number of analyzed individuals was small, it is important to highlight that we found other variants in the number of copies that have been described in association with neurodevelopmental disorders, congenital anomalies, deafness, and short/tall stature, among others, in almost half of them, which will probably impact the phenotype negatively in patients with aneuploidies.
Downloads
References
Hutaff-Lee C, Cordeiro L, Tartaglia N. Cognitive and medical features of chromosomal aneuploidy. Handb Clin Neurol. 2013;111:273‑9. https://doi.org/10.1016/B978-0-444-52891-9.00030-0
Nielsen J, Holm V, Haahr J. Prevalence of Edwards’ syndrome. Clustering and seasonal variation? Humangenetik. 1975;26:113‑6. https://doi.org/10.1007/BF00278437
Savva GM, Walker K, Morris JK. The maternal age-specific live birth prevalence of trisomies 13 and 18 compared to trisomy 21 (Down syndrome). Prenat Diagn. 2010;30:57‑64. https://doi.org/10.1002/pd.2403
Sherman SL, Allen EG, Bean LH, Freeman SB. Epidemiology of Down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13:221‑7. https://doi.org/10.1002/mrdd.20157
Abarca-Barriga HH, Chávez-Pastor MA, Trubnykova M, Vásquez F, Poterico JA. Chromosomal microarray analysis in Peruvian children with delayed psychomotor development or intellectual disability. Rev Peru Med Exp Salud Pública. 2017;34:572‑4. https://doi.org/10.17843/rpmesp.2017.343.2741
de Smith AJ, Trewick AL, Blakemore AIF. Implications of copy number variation in people with chromosomal abnormalities: Potential for greater variation in copy number state may contribute to variability of phenotype. Hugo J. 2010;4:1‑9. https://doi.org/10.1007/s11568-010-9144-z
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848‑53. https://doi.org/10.1126/science.1136678
Henrichsen CN, Vinckenbosch N, Zöllner S, Chaignat E, Pradervand S, Schütz F, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424‑9. https://doi.org/10.1038/ng.345
Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176‑87. https://doi.org/10.1093/hmg/ddq366
Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15:478‑81. https://doi.org/10.1038/gim.2012.164
Schiaffino MC, Bellini C, Costabello L, Caruso U, Jakobs C, Salomons GS, et al. X-linked creatine transporter deficiency: Clinical description of a patient with a novel SLC6A8 gene mutation. Neurogenetics. 2005;6:165‑8. https://doi.org/10.1007/s10048-005-0002-4
Rosenberg C, Freitas ÉL, Uehara DT, Auricchio MTBM, Costa SS, Oiticica J, et al. Genomic copy number alterations in non-syndromic hearing loss. Clin Genet. 2016;89:473‑7. https://doi.org/10.1111/cge.12683
Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, et al. 3q29 microdeletion syndrome: Clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154‑60. https://doi.org/10.1086/431653
Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: Case report and review. Am J Med Genet A. 2010;152A:2459‑67. https://doi.org/10.1002/ajmg.a.33573
Carroll LS, Williams HJ, Walters J, Kirov G, O’Donovan MC, Owen MJ. Mutation screening of the 3q29 microdeletion syndrome candidate genes DLG1 and PAK2 in schizophrenia. Am J Med Genet Part B Neuropsychiatr Genet. 2011;156B:844‑9. https://doi.org/10.1002/ajmg.b.31231
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu T, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838‑46. https://doi.org/10.1038/ng.909
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216‑21. https://doi.org/10.1038/nature13908
Warnica W, Merico D, Costain G, Alfred SE, Wei J, Marshall CR, et al. Copy number variable microRNAs in schizophrenia and their neurodevelopmental gene targets. Biol Psychiatry. 2015;77:158‑66. https://doi.org/10.1016/j.biopsych.2014.05.011
Zeng Y, Broxmeyer HE, Staser K, Chitteti BR, Park S-J, Hahn S, et al. Pak2 regulates hematopoietic progenitor cell proliferation, survival, and differentiation. Stem Cells Dayt Ohio. 2015;33:1630‑41. https://doi.org/10.1002/stem.1951
Wang Y, Zeng C, Li J, Zhou Z, Ju X, Xia S, et al. PAK2 haploinsufficiency results in synaptic cytoskeleton impairment and autism-related behavior. Cell Rep. 2018;24:2029‑41. https://doi.org/10.1016/j.celrep.2018.07.061
Singh H, Tiwari P, Bhavi V, Chaudhary PS, Suravajhala P, Mohan MK, et al. Application of chromosomal microarray for evaluation of idiopathic short stature in Asian Indian children: A pilot study. Indian J Endocrinol Metab. 2018;22:100‑6. https://doi.org/10.4103/ijem.IJEM_202_17
Meier N, Bruder E, Lapaire O, Hoesli I, Kang A, Hench J, et al. Exome sequencing of fetal anomaly syndromes: Novel phenotype–genotype discoveries. Eur J Hum Genet. 2019;27:730‑7. https://doi.org/10.1038/s41431-018-0324-y
DECIPHER v9.7: Mapping the clinical genome. Fecha de consulta: 20 mayo de 2016. Disponible en: https://decipher.sanger.ac.uk/browser
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524‑33. https://doi.org/10.1016/j.ajhg.2009.03.010
Sailani MR, Makrythanasis P, Valsesia A, Santoni FA, Deutsch S, Popadin K, et al. The complex SNP and CNV genetic architecture of the increased risk of congenital heart defects in Down syndrome. Genome Res. 2013;23:1410‑21. https://doi.org/10.1101/gr.147991.112
Rambo-Martin BL, Mulle JG, Cutler DJ, Bean LJH, Rosser TC, Dooley KJ, et al. Analysis of copy number variants on chromosome 21 in Down syndrome-associated congenital heart defects. G3 (Bethesda). 2017;8:105‑11. https://doi.org/10.1534/g3.117.300366
Mégarbané A, Noguier F, Stora S, Manchon L, Mircher C, Bruno R, et al. The intellectual disability of trisomy 21: Differences in gene expression in a case series of patients with lower and higher IQ. Eur J Hum Genet. 2013;21:1253‑9. https://doi.org/10.1038/ejhg.2013.24
Park J, Chung KC. New perspectives of Dyrk1A role in neurogenesis and neuropathologic features of Down syndrome. Exp Neurobiol. 2013;22:244‑8. https://doi.org/10.5607/en.2013.22.4.244
Rodríguez-Cadilla MR. Consideraciones bioéticas y jurídicas de la información genética y el diagnóstico prenatal. Vox Juris. 2015;28:15‑40.
Abarca-Barriga H. Perfil epidemiológico de las anomalías genéticas y congénitas en el Servicio de Citogenética y Citopatología del Hospital Nacional Guillermo Almenara Irigoyen (tesis). Lima: Universidad Nacional Mayor de San Marcos; 2007.
Carreira IM, Ferreira SI, Matoso E, Pires LM, Ferrão J, Jardim A, et al. Copy number variants prioritization after array-CGH analysis – a cohort of 1000 patients. Mol Cytogenet. 2015;8:103‑11. https://doi.org/10.1186/s13039-015-0202-z
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2019;22:245-57. https://doi.org/10.1038/s41436-019-0686-8
Fan Y-S, Ouyang X, Peng J, Sacharow S, Tekin M, Barbouth D, et al. Frequent detection of parental consanguinity in children with developmental disorders by a combined CGH and SNP microarray. Mol Cytogenet. 2013;6:38‑43. https://doi.org/10.1186/1755-8166-6-38
Rinaldi B, van Hoof E, Corveleyn A, van Cauter A, de Ravel T. BCAP31-related syndrome: The first de novo report. Eur J Med Genet. 2019;103732. https://doi.org/10.1016/j.ejmg.2019.103732
Sayee R, Thomas IM. Consanguinity, non-disjunction, parental age and Down’s syndrome. J Indian Med Assoc. 1998;96:335‑7.
Some similar items:
- Juan Carlos Herrera, Luis Fernando Isaza, José Luis Ramírez, Gonzalo Vásquez, Carlos Mario Muñetón, Detection of chromosome 17 aneuplody and TP53 gene deletion in a broad variety of solid tumors by dual-color fluorescence in situ hybridization (FISH) , Biomedica: Vol. 30 No. 3 (2010)
- Marita Lardón, José Uberos, Eduardo Narbona, Does corticosteroid treatment during the pre and postnatal periods affect the neurodevelopmental outcome of premature newborns? , Biomedica: Vol. 37 No. Sup.1 (2017): Suplemento 1, Alteraciones del sistema nervioso
- Catalina Arango-Ferreira, Diana Isabel Villegas, Laura Daniela Burbano, Augusto Quevedo, Follow up of HIV perinatal exposure and accomplishment of strategies to reduce the risk of viral transmission, experience in a reference hospital in Medellín , Biomedica: Vol. 39 No. Supl. 2 (2019): Enfermedades transmisibles en el trópico, agosto
- Katherine Barrios , Oscar Patiño, Nelson Muñoz, Carlos Moneriz, Congenital Langerhans cell histiocytosis , Biomedica: Vol. 40 No. 3 (2020)
- Lorena Peñaloza, Catalina Forero, Camila Céspedes, Characterization of patients diagnosed with congenital hypothyroidism at the Hospital Universitario San Ignacio between 2001 and 2017 , Biomedica: Vol. 40 No. 3 (2020)
- Hernando Baquero, María Elena Venegas Martinez, Lorena Velandia Forero, Fredy Neira Safi, Edgar Navarro, Neonatal late-onset infection with SARS CoV-2 , Biomedica: Vol. 40 No. Supl. 2 (2020): SARS-CoV-2 y COVID-19
- Sergio Iván Agudelo, Carlos Federico Molina, Óscar Andrés Gamboa, Juan David Suárez, Direct costs of neonatal infection acquired in the community in full-term newborns and low risk at birth, Cundinamarca, Colombia , Biomedica: Vol. 41 No. 1 (2021)
- Katherine Márquez, Diego Andrés Rodríguez, Luis Alfonso Pérez, Mauricio Duarte, Luis Augusto Zárate, Epidermolysis bullosa with pyloric atresia: Report of two cases in consecutive siblings , Biomedica: Vol. 41 No. 2 (2021)
- María Fernanda Unigarro, Catalina Forero , Camila Céspedes , Neuropsychological and physical development of patients diagnosed with congenital hypothyroidism at the San Ignacio University Hospital between 2001 and 2017 , Biomedica: Vol. 42 No. Sp. 1 (2022): Mayo, Enfermedades crónicas en el trópico
- Diana Ramírez , Víctor Ramos , Sandra Navarro, Adriana Montealegre, Julia Arciniegas , X-ray radiation dose and associated factors in neonates from the newborn unit of Hospital Universitario San Ignacio, Bogotá, Colombia , Biomedica: Vol. 43 No. 3 (2023)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























