Molecular and phenotypic characterization of Salmonella Typhimurium monophasic variant (1,4,[5],12:i:-) from Colombian clinical isolates
Abstract
Introduction. The Salmonella Typhimurium monophasic variant (1,4,[5],12:i:-) is currently the most commonly detected variant in Salmonella surveillance programs worldwide. In Colombia, the Salmonella enterica monophasic variant is the fourth most common clinical isolate recovered through the laboratory surveillance of the Grupo de Microbiología from the Instituto Nacional de Salud; however, it is unknown whether these isolates are closely related to the monophasic Typhimurium variant, which circulates globally, and their genetic and phenotypic characteristics have not been reported.
Objective. To characterize monophasic Salmonella enterica isolates identified in Colombia from 2015 to 2018 by the Instituto Nacional de Salud.
Materials and methods. Two hundred eighty-six clinical isolates of the monophasic Salmonella enterica variant were analyzed by PCR or whole-genome sequencing to confirm whether they corresponded to the Salmonella Typhimurium monophasic variant while the genetic structure of the operon encoding the second flagellar phase was determined in 54 isolates. Motility, growth, and expression of the outer membrane proteins were evaluated in 23 isolates.
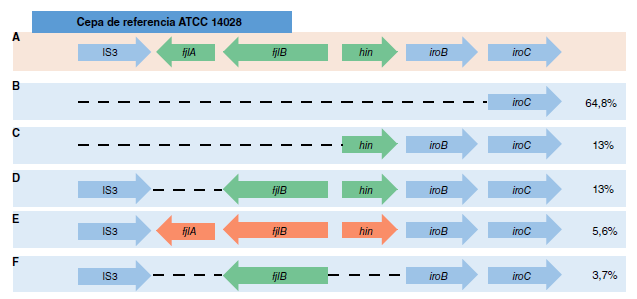
Results. During the study period in Colombia, 61% (n=174) of Salmonella monophasic isolates belonged to Salmonella Typhimurium serovar monophasic (1,4,[5],12:i-). Of these, 64.8% (n=35/54) were related to the European/Spanish clone and 13% (n=7/54) to the U.S. clone. Two isolates recovered from urine samples showed differences in motility, growth, and the absence of the OmpD porin in M9 minimal medium.
Conclusions. Most of the monophasic Salmonella Typhimurium variants that have circulated in Colombia since 2015 lacked the second phase of operon fljAB, which is related to the European/Spanish clone. The results evidenced phenotypic changes in urine samples suggesting bacterial adaptation in the case of these invasive samples.
Downloads
References
World Health Organization. Salmonella (no tifoidea). Fecha de consulta: 27 de abril de 2019. Disponible en: https://www.who.int/es/news-room/fact-sheets/detail/salmonella-(nontyphoidal) 2. Grimont PA, Weill FX. Antigenic formulae of the Salmonella serovars. 9th edition. Paris: Institut Pasteur; 2007. p. 166.
Yamamoto S, Kutsukake K. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:958-67. https://doi.org/10.1128/JB.188.3.958-967.2006
Bonifield HR, Hughes KT. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol. 2003;185:3567-74. https://doi.org/10.1128/JB.185.12.3567-3574.2003
Centers for Disease Control and Prevention (CDC). National Enteric Disease Surveillance. Salmonella Annual Report 2016. Atlanta, USA: CDC; 2018.
European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA. 2019;17:e05926.
https://doi.org/10.2903/j.efsa.2019.5926
Arai N, Sekizuka T, Tamamura Y, Tanaka K, Barco L, Izumiya H, et al. Phylogenetic characterization of Salmonella enterica serovar Typhimurium and its monophasic variant isolated from food animals in japan revealed replacement of major epidemic clones in the last 4 decades. J Clin Microbiol. 2018;56:e01758-17. https://doi.org/10.1128/JCM.01758-17
Mastrorilli E, Pietrucci D, Barco L, Ammendola S, Petrin S, Longo A, et al. A comparative genomic analysis provides novel insights into the ecological success of the monophasic salmonella serovar 4,[5],12:i:. Front Microbiol. 2018;9:715. https://doi.org/10.3389/fmicb.2018.00715
Sun H, Wan Y, Du P, Bai L. The epidemiology of monophasic Salmonella typhimurium. Foodborne Pathog Dis 2020;17:87-97. https://doi.org/10.1089/fpd.2019.2676
Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, et al. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl Environ Microbiol. 2010;76:4601-10. https://doi.org/10.1128/AEM.02991-09
Soyer Y, Moreno-Switt A, Davis MA, Maurer J, McDonough PL, Schoonmaker-Bopp DJ, et al. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J Clin Microbiol. 2009;47:3546-56. https://doi.org/10.1128/JCM.00546-09
Barco L, Longo A, Lettini AA, Cortini E, Saccardin C, Minorello C, et al. Molecular characterization of “inconsistent” variants of Salmonella Typhimurium isolated in Italy. Foodborne Pathog Dis. 2014;11:497-9. https://doi.org/10.1089/fpd.2013.1714
Rodríguez EC, Díaz-Guevara P, Moreno J, Bautista A, Montaño L, Realpe ME, et al. Laboratory surveillance of Salmonella enterica from human clinical cases in Colombia 2005-2011. Enferm Infecc Microbiol Clin. 2017;35:417-25. https://doi.org/10.1016/j.eimc.2016.02.023
Instituto Nacional de Salud. Vigilancia por laboratorio de Salmonella spp. [Internet]. Bogotá: Instituto Nacional de Salud; 2018.
Li Y, Pulford CV, Díaz P, Pérez-Sepúlveda BM, Duarte C, Predeus AV, et al. Genomic and phylogenetic analysis of Salmonella Typhimurium and its monophasic variants responsible for invasive endemic infections in Colombia. BioRxiv. 2019. https://doi.org/10.1101/588608
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 27th edition. CLSI supplement M100. Wayne, PA: CLSI; 2017.
Green MR, Michael R, Sambrook J. Molecular cloning: A laboratory manual. 4th edition. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2014.
Pérez-Sepúlveda BM, Heavens D, Pulford CV, Predeus AV, Low R, Webster H, et al. An accessible, efficient and global approach for the large-scale sequencing of bacterial genomes. BioRxiv. 2020. https://doi.org/10.1101/2020.07.22.200840
McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852-6. https://doi.org/10.1038/35101614
Echeita MA, Herrera S, Usera MA. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J Clin Microbiol. 2001;39:2981-3. https://doi.org/10.1128/JCM.39.8.2981-2983.2001
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535-42. https://doi.org/10.1093/nar/gkw1017
Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, et al. Identification of novel factors involved in modulating motility of Salmonella enterica serotype typhimurium. PLoS ONE. 2014;9:e111513. https://doi.org/10.1371/journal.pone.0111513
Villarreal JM, Becerra-Lobato N, Rebollar-Flores JE, Medina-Aparicio L, Carbajal-GómezE, Zavala-García ML, et al. The Salmonella enterica serovar Typhi ltrR-ompR-ompC-ompF genes are involved in resistance to the bile salt sodium deoxycholate and in bacterial transformation. Mol Microbiol. 2014;92:1005-24. https://doi.org/10.1111/mmi.12610
Echeita-Sarrionandia MA, León SH, Baamonde CS. Invasive gastroenteritis, anything new? Enferm Infecc Microbiol Clin. 2011;29(Suppl.3):55-60. https://doi.org/10.1016/S0213-005X(11)70029-5
Hopkins KL, Kirchner M, Guerra B, Granier SA, Lucarelli C, Porrero MC, et al. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: A new pandemic strain? Euro Surveill. 2010;15:19580.
Cito F, Baldinelli F, Calistri P, Di Giannatale E, Scavia G, Orsini M, et al. Outbreak of unusual Salmonella enterica serovar Typhimurium monophasic variant 1,4 [5],12:i:-, Italy, June 2013 to September 2014. Euro Surveill. 2016;21. https://doi.org/10.2807/1560-7917.ES.2016.21.15.30194
European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017;15. https://doi.org/10.2903/j.efsa.2017.5077
Machado J, Bernardo F. Prevalence of Salmonella in chicken carcasses in Portugal. J Appl Bacteriol 1990;69:477-80. https://doi.org/10.1111/j.1365-2672.1990.tb01538.x
Echeita MA, Aladueña A, Cruchaga S, Usera MA. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:- strain in Spain. J Clin Microbiol. 1999;37:3425. https://doi.org/10.1128/JCM.37.10.3425-3425.1999
Helmuth IG, Espenhain L, Ethelberg S, Jensen T, Kjeldgaard J, Litrup E, et al. An outbreak of monophasic Salmonella Typhimurium associated with raw pork sausage and other pork products, Denmark 2018-19. Epidemiol Infect. 2019;147:e315. https://doi.org/10.1017/S0950268819002073
Magossi G, Bai J, Cernicchiaro N, Jones C, Porter E, Trinetta V. Seasonal presence of Salmonella spp., Salmonella Typhimurium and its monophasic variant serotype I 4,[5],12:i:-, in selected United States Swine Feed Mills. Foodborne Pathog Dis 2019;16:276-81. https://doi.org/10.1089/fpd.2018.2504
Yang X, Wu Q, Zhang J, Huang J, Guo W, Cai S. Prevalence and characterization of monophasic Salmonella serovar 1,4,[5],12:i:- of food origin in China. PLoS ONE. 2015;10:e0137967. https://doi.org/10.1371/journal.pone.0137967
Mulvey MR, Finley R, Allen V, Ang L, Bekal S, El Bailey S, et al. Emergence of multidrugresistant Salmonella enterica serotype 4,[5],12:i:- involving human cases in Canada: Results from the Canadian Integrated Program on Antimicrobial Resistance Surveillance (CIPARS), 2003-10. J Antimicrob Chemother. 2013;68:1982-6. https://doi.org/10.1093/jac/dkt149
Tavechio AT, Fernandes SA, Ghilardi ÂC, Soule G, Ahmed R, Melles CE. Tracing lineage by phenotypic and genotypic markers in Salmonella enterica subsp. enterica serovar 1,4,[5],12:i:- and Salmonella Typhimurium isolated in state of São Paulo, Brazil. Mem Inst Oswaldo Cruz. 2009;104:1042-6. https://doi.org/10.1590/S0074-02762009000700019
Vico JP, Lorenzutti AM, Zogbi AP, Aleu G, Sánchez IC, Caffer MI, et al. Prevalence, associated risk factors, and antimicrobial resistance profiles of non-typhoidal Salmonella in large scale swine production in Córdoba, Argentina. Res Vet Sci. 2020;130:161-9. https://doi.org/10.1016/j.rvsc.2020.03.003
Elnekave E, Hong S, Mather AE, Boxrud D, Taylor AJ, Lappi V, et al. Salmonella enterica serotype 4,[5],12:i:- in Swine in the United States Midwest: An emerging multidrug-resistant clade. Clin Infect Dis. 2018;66:877-85. https://doi.org/10.1093/cid/cix909
de la Torre E, Zapata D, Tello M, Mejía W, Frías N, García-Peña FJ, et al. Several Salmonella enterica subsp. enterica serotype 4,5,12:i:- phage types isolated from swine samples originate from serotype typhimurium DT U302. J Clin Microbiol. 2003;41:2395-400. https://doi.org/10.1128/jcm.41.6.2395-2400.2003
Donado-Godoy P, Clavijo V, León M, Arévalo A, Castellanos R, Bernal J, et al. Counts, serovars, and antimicrobial resistance phenotypes of Salmonella on raw chicken meat at retail in Colombia. J Food Prot. 2014;77:227-35. https://doi.org/10.4315/0362-028X.JFP-13-276
Rodríguez JM, Rondón IS, Verjan N. Serotypes of Salmonella in broiler carcasses marketed at Ibagué, Colombia. Rev Bras Cienc Avic. 2015;17:545-52. https://doi.org/10.1590/1516-635X1704545-552
Rondón-Barragán IS, Arcos EC, Mora-Cardona L, Fandiño C. Characterization of Salmonella species from pork meat in Tolima, Colombia. Revista Colombiana de Ciencias Pecuarias. 2015;28:74-82.
Durango J, Arrieta G, Mattar S. Presencia de Salmonella spp. en un área del Caribe colombiano: un riesgo para la salud pública. Biomédica. 2004;24:89-96. https://doi.org/10.7705/biomedica.v24i1.1252
Petrovska L, Mather AE, AbuOun M, Branchu P, Harris SR, Connor T, et al. Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005-2010. Emerging Infect Dis. 2016;22:617-24. https://doi.org/10.3201/eid2204.150531
Crayford G, Coombes JL, Humphrey TJ, Wigley P. Monophasic expression of FliC gen by Salmonella 4,[5],12:i:- DT193 does not alter its pathogenicity during infection of porcine intestinal epithelial cells. Microbiology. 2014;160:2507-16. https://doi.org/10.1099/mic.0.081349-0
Tena D, González-Praetorius A, Pérez-Pomata MT, Gimeno C, Alén MJ, Robres P, et al. Urinary infection caused by non typhi Salmonella. Enferm Infecc Microbiol Clin. 2000;18:79-82.
Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103-18. https://doi.org/10.1046/j.1365-2958.2003.03313.x
Ipinza F, Collao B, Monsalva D, Bustamante VH, Luraschi R, Alegría-Arcos M, et al. Participation of the Salmonella OmpD porin in the infection of RAW264.7 macrophages and BALB/c mice. PLoS ONE 2014;9:e111062. https://doi.org/10.1371/journal.pone.0111062
Some similar items:
- Katherine Laiton-Donato, José A. Usme-Ciro, Angélica Rico, Lissethe Pardo, Camilo Martínez, Daniela Salas, Susanne Ardila, Andrés Páez, Phylogenetic analysis of Chikungunya virus in Colombia: Evidence of purifying selection in the E1 gene , Biomedica: Vol. 36 (2016): Suplemento 2, Enfermedades virales
- José Joaquín Carvajal, Nildimar Alves Honorio, Silvia Patricia Díaz, Edinso Rafael Ruiz, Jimmy Asprilla, Susanne Ardila, Gabriel Parra-Henao, Detection of Aedes albopictus (Skuse) (Diptera: Culicidae) in the municipality of Istmina, Chocó, Colombia , Biomedica: Vol. 36 No. 3 (2016)
- Nélida Muñoz, María Elena Realpe, Elizabeth Castañeda, Clara Inés Agudelo, Characterization by pulsed-field gel electrophoresis of Salmonella Typhimurium isolates recovered in the acute diarrheal disease surveillance program in Colombia, 1997-2004 , Biomedica: Vol. 26 No. 3 (2006)
- Myrtha Arango, Elizabeth Castañeda, Clara Inés Agudelo, Catalina De Bedout, Carlos Andrés Agudelo, Angela Tobón, Melva Linares, Yorlady Valencia, Ángela Restrepo, The Colombian Histoplasmosis Study Group, Histoplasmosis: results of the Colombian National Survey, 1992-2008 , Biomedica: Vol. 31 No. 3 (2011)
- Raúl Murillo, Ricardo Cendales, Carolina Wiesner, Marion Piñeros, Sandra Tovar, Effectiveness of cytology-based cervical cancer screening in the Colombian health system , Biomedica: Vol. 29 No. 3 (2009)
- Sandra Lorena Girón, Julio César Mateus, Fabián Méndez, Impact of an open waste disposal site on the occurrence of respiratory symptoms and on health care costs of children , Biomedica: Vol. 29 No. 3 (2009)
- Jaime E. Bernal, Martha Lucía Tamayo , Ignacio Briceño , Escilda Benavides , Newborn screening in Colombia: The experience of a private program in Bogotá , Biomedica: Vol. 44 No. 1 (2024)
- Andrés Páez, Gloria Rey, Carlos Agudelo, Alvaro Dulce, Edgar Parra, Hernando Díaz-Granados, Damaris Heredia, Luis Polo, Outbreak of urban rabies transmitted by dogs in Santa Marta, northern Colombia , Biomedica: Vol. 29 No. 3 (2009)
- Patricia Escobar, Katherine Paola Luna, Indira Paola Hernández, César Mauricio Rueda, María Magdalena Zorro, Simon L. Croft, In vitro susceptibility of Trypanosoma cruzi strains from Santander, Colombia, to hexadecylphosphocholine (miltefosine), nifurtimox and benznidazole , Biomedica: Vol. 29 No. 3 (2009)
- Mauricio Beltrán, María Cristina Navas, María Patricia Arbeláez, Jorge Donado, Sergio Jaramillo, Fernando De la Hoz, Cecilia Estrada, Lucía del Pilar Cortés, Amalia de Maldonado, Gloria Rey, Seroprevalence of hepatitis B virus and human immunodeficiency virus infection in a population of multiply-transfused patients in Colombia , Biomedica: Vol. 29 No. 2 (2009)
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |

























